Abstract
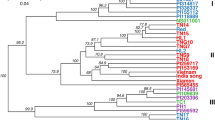
Radix bupleuri, is an important traditional Chinese medicine resource. The lack of genetic molecular markers of Radix bupleuri turned to be an obstacle for genetic analysis and accessions identification. In this study, SNP loci were detected based on genotyping-by-sequencing (GBS) and the genetic diversity of 39 Bupleurum varieties were examined. A total of 25.1 Gb of data was obtained by sequencing, with an average of 0.64 Gb per sample. After screening, 83 898 high-quality SNPs were obtained. The 39 accessions could be divided into three major groups based on population structure analysis, Neighbor-joining clustering and principal component analysis. The average observed heterozygosity and expected heterozygosity of Bupleurum populations were 0.24 and 0.17, respectively, indicating that Bupleurum populations from five different provinces had a low level of genetic diversity. Population nucleotide diversity analysis and analysis of molecular variance showed that the percentage of intrapopulation variation was 92.13%, while the percentage of interpopulation variation was only 7.87%. There was relative aggregation of Bupleurum samples with the same geographical origin, but the division of population structure was not completely correlated with sample origin. The results showed that the genetic diversity of the materials was narrow. This provides a good basis for the genetic breeding and protection of species diversity of Bupleurum.



Similar content being viewed by others
REFERENCES
Feng, Y.J., Wu, Z.W., Luo, Y.Y., et al., A new triterpene diglycoside from the roots of Bupleurum chinense DC. and its inhibitory effect on adipogensis in 3T3-L1 cells, Med. Chem. Res., 2019, vol. 28, pp. 239—245. https://doi.org/10.1007/s00044-018-2279-5
Pan, S.L., Bupleurum Species: Scientific Evaluation and Clinical Applications, Boca Raton: CRC Press, 2006.
Law, Y.K., Mo, J.F., Wong, K.W., Autophagic effects of Chaihu (dried roots of Bupleurum chinense DC or Bupleurum scorzoneraefolium Wild), Chinese Med., 2014, 9:21. vol. 9, no. 1. https://doi.org/10.1186/1749-8546-9-21.
Yu, J., Deng, A., Wu, L., et al., Osteoclast-inhibiting saikosaponin derivatives from Bupleurum Chinense, Fitoterapia, 2013, vol. 85, pp. 101—108. https://doi.org/10.1016/j.fitote.2013.01.005
Li, D.Q., Wu, J., Liu, L.Y., et al., Cytotoxic triterpenoid glycosides (saikosaponins) from the roots of Bupleurum chinense, Bioorg. Med. Chem. Lett., 2015, vol. 25, no. 18, pp. 3887—3892. https://doi.org/10.1016/j.bmcl.2015.07.053
Li, H.Y., Zhao, Y.H., Zeng, M.J., et al., Saikosaponin D relieves unpredictable chronic mild stress induced depressive-like behavior in rats: involvement of HPA axis and hippocampal neurogenesis, Psychopharmacology, 2017, vol. 234, pp. 3385—3394. https://doi.org/10.1007/s00213-017-4720-8
Wang, Y., Qiang, G., Cheng, Z., et al., New saikosaponins from the roots of Bupleurum chinense, Phytochem. Lett., 2017, vol. 21, pp. 183—189. https://doi.org/10.1016/j.phytol.2017.06.005
Wang, H.W., Liu, M., Zhong, T.D., et al., Saikosaponin-d attenuates ventilator-induced lung injury in rats, Int. J. Clin. Exp. Med., 2015, vol. 8, no. 9, pp. 15137—15145. http://www.ijcem.com/ISSN:1940-5901/IJCEM0004264.
Chen, X.Q., Chen, S.J., Liang, W.N., et al., Saikosaponin A attenuates perimenopausal depression-like symptoms by chronic unpredictable mild stress, Neurosci. Lett., 2018, vol. 662, pp. 283—289. https://doi.org/10.1016/j.neulet.2017.09.046
Lorrai, I., Maccioni, P., Carai, M., et al., Suppressing effect of saikosaponin A, an active ingredient of Bupleurum falcatum, on chocolate self-administration and reinstatement of chocolate seeking in rats, Neurosci. Lett., 2017, vol. 638, pp. 211—217. https://doi.org/10.1016/j.neulet.2016.12.019
National Pharmacopoeia Committee, Pharmacopoeia of Peoples Republic of China, Beijing: Chemical Industry Press, 2020, part 1, appendix 2, p. 293.
Sui, C., He, W.J., Lin, C.S., et al., Development of genomic SSR and potential EST-SSR markers in Bupleurum chinense DC., Afr. J. Biotechnol., 2009, vol. 8, no. 22, pp. 6233—6240. https://doi.org/10.4314/ajb.v8i22.66126
Zhao, X., Liu, C., Xue, W., et al., ISSR research on germplasm of Bupleurum chinense DC. in Beijing, Mod. Chin. Med., 2015, vol. 17, no. 10, pp. 1008—1013. https://doi.org/10.13313/j.issn.1673-4890.2015.10.004
Yang, W., Bai, Y., and Hu, J.Y., ISSR research on germplasm of Bupleurum chinense DC.in Baokang, Chin. Med. J. Res. Pract., 2013, vol. 27, no. 2, pp. 25—27. https://doi.org/10.13728/j.1673-6427.2013.02.008
Lee, K.J., Lee, J.R., Sebasti, N.R., et al., Genetic diversity assessed by genotyping by sequencing (GBS) in watermelon germplasm, Genes, 2019, vol. 10, pp. 822—834. https://doi.org/10.3390/genes10100822
Fu, Y.B. and Peterson, G.W., Genetic diversity analysis with 454 pyrosequencing and genomic reduction confirmed the eastern and western division in the cultivated barley gene pool, Plant Genome, 2011, vol. 4, no. 3, pp. 226—237. https://doi.org/10.3835/plantgenome2011.08.0022
Peterson, B.K., Weber, J.N., Kay, E.H., et al., Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species, PLoS One, 2012, vol. 7. e37135. https://doi.org/10.1371/journal.pone.0037135
Peterson, G., Dong, Y., Horbach, C., et al., Genotyping-by-sequencing for plant genetic diversity analysis: a lab guide for SNP genotyping, Diversity, 2014, vol. 6, no. 4, pp. 665—680. https://doi.org/10.3390/d6040665
Yu, D., Wang, H., Gu, W., et al., Genetic diversity and population structure of popcorn germplasm resources using genome-wide SNPs through genotyping-by-sequencing, Genet. Resour. Crop Evol., 2021, vol. 68, pp. 2379—2389. https://doi.org/10.1007/s10722-021-01137-0
Lee, H.Y., Jang, S., Yu, C.R., et al., Population structure and genetic diversity of Cucurbita moschata based on genome-wide high-quality SNPs, Plants, 2020, vol. 10, no. 1, pp. 56—65. https://doi.org/10.3390/plants10010056
Yang, X., Tan, B., Liu, H., et al., Genetic diversity and population structure of Asian and European common wheat accessions based on genotyping-by-sequencing, Front. Genet., 2020, vol. 11. e580782. https://doi.org/10.21203/rs.2.17640/v1
Li, H. and Durbin, R., Fast and accurate short read alignment with Burrows—Wheeler transform, Bioinformatics, 2009, vol. 25, no. 14, pp. 1754—1760. https://doi.org/10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., et al., The sequence alignment/map format and SAMtools, Bioinformatics, 2009, vol. 25, no. 16, pp. 2078—2079. https://doi.org/10.1093/bioinformatics/btp352
Wang, K., Li, M., and Hakonarson, H., ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data, Nucleic Acids Res., 2010, vol. 38, no. 16. e164. https://doi.org/10.1093/nar/gkq603
Alexander, D.H., Novembre, J., and Lange, K., Fast model-based estimation of ancestry in unrelated individuals, Genome. Res., 2009, vol. 19, pp. 1655—1664. https://doi.org/10.1101/gr.094052.109
Excoffier, L., Laval, G., and Schneider, S., Arlequin (version 3.0): an integrated software package for population genetics data analysis, Evol. Bioinf. Online, 2005, vol. 1, pp. 47—50. https://doi.org/10.1143/JJAP.34.L418
Nei, M., Estimation of average heterozygosity and genetic distance from a small number of individuals, Genetics, 1978, vol. 89, no. 3, pp. 583—590. https://doi.org/10.1007/BF00155576
World Health Organization, WHO Monographs on Selected Medicinal Plants, Geneva: World Health Organization, 1999, vol. 1.
Huang, H.Q., Zhang, X., Xu, Z.X., et al., Fast determination of saikosaponins in Bupleurum by rapid resolution liquid chromatography with evaporative light scattering detection. J. Pharm. Biomed. Anal., 2009, vol. 49, no. 4, pp. 1048—1055. https://doi.org/10.1016/j.jpba.2009.01.011
Huang, W., Sun, P., Zhang, W.S., et al., Genetic diversity of Bupleurum chinense DC. populations from different altitudes in Dongling mountain district in Beijing, Plant Genet. Resour., 2008, vol. 9, no. 4, pp. 453—457.
Ke, S.Y., Shi, L.L., Ma, Y.Z., et al., Evaluation of the genetic diversity of Bupleurum using amplified fragment length polymorphism analysis, Genet. Mol. Res., 2015, vol. 14, no. 1, pp. 2590—2599. https://doi.org/10.4238/2015.March.30.18
Du, S.M., Wang, G., Liu, Y.M., et al., Study on biological materials with genetic diversity of Bupleurum marginatum in Northwest of Hubei province of China based on ISSR, Adv. Mat. Res., 2013, vol. 830, pp. 463—468. https://doi.org/10.4028/www.scientific.net/AMR.830.463
Li, Y.H., Yu, X.L., Ou, X.J., et al., Genetic diversity of different origin Bupleurum chinese detected by ISSR analysis, Lishizhen Med. Mater. Med. Res., 2018, vol. 29, pp. 1728—1731.
ACKNOWLEDGMENTS
We would like to thank Springer Nature for providing linguistic assistance for manuscript preparation.
Funding
This work was supported by the Key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302), the State Key Laboratory of Pharmaceutical New-tech for Chinese Medicine (SKL2020M0302) and the Talent training project funded by the central government to support the reform and development of local colleges and Universities (ZYRCB2021008).
Author information
Authors and Affiliations
Contributions
M. Jiang, S. Yan, W.C. Ren, and N.N. Xing contributed equally to this work.
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This aitical does not contain any experiment with human participants or animals.
Rights and permissions
About this article
Cite this article
Jiang, M., Yan, S., Ren, W.C. et al. Genetic Diversity and Population Structure of Traditional Chinese Herb Radix bupleuri Resources Using Genome-Wide SNPs through Genotyping-by-Sequencing. Russ J Genet 58, 1485–1492 (2022). https://doi.org/10.1134/S1022795422120055
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795422120055