Abstract
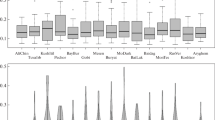
A decrease in the level of genetic diversity is one of the main genetic problems in pig breeding worldwide. In Russia, some local breeds are endangered because of their low number. A strong directional selection can lead to a decrease in the diversity in transboundary breeds. In this regard, the aim of our study was to estimate the genetic diversity and to establish phylogenetic relationships in nine pig breeds reared in Russia by the analysis of mtDNA D-loop sequences. In total, 273 nucleotide sequences of mtDNA D-loop were sequenced in pigs of the following breeds: Breit, Kemerovo, Livni, Murom, Urzhum, Mangalitsa, Large White, Landrace, and Duroc. In the entire sample, 84 haplotypes, including 55 in local and 29 in transboundary breeds, and 104 variable sites were identified. Local breeds were characterized by a lower average number of nucleotide differences between the haplotypes than transboundary ones (K = 6.272 and K = 9.934). Of the studied pigs, 78.8% belonged to the haplogroup E; haplogroups D (20.5%) and A (0.7%) were found less frequently. The AMOVA analysis demonstrated that 43.88% of total genetic variability was due to the differences between the studied breeds. The analysis of the median network structure demonstrated that the Kemerovo breed was the most differentiated among local breeds, while the Mangalitsa breed entered the cluster of transboundary breeds owing to its European origin. The results obtained can be useful monitoring the genetic diversity in transboundary breeds, as well as for developing evidence-based programs for local pig breed conservation.




Similar content being viewed by others
REFERENCES
Song, R., Wang, Yu., Wang, Ya., and Zhao, J., Base editing in pigs for precision breeding, Front. Agr. Sci. Eng., 2020, vol. 7, no. 2, pp. 161—170. https://doi.org/10.15302/J-FASE-2019308
Park, H.-S., Min, B., and Oh, S.-H., Research trends in outdoor pig production—a review, Asian-Australas. J. Anim. Sci., 2017, vol. 30, no. 9, pp. 1207—1214. https://doi.org/10.5713/ajas.17.0330
Thi, L., Huyen, T., Roessler, R., et al., Impact of the Use of Exotic Compared to Local Pig Breeds on Socio-Economic Development and Biodiversity in Vietnam, Stuttgart: Grauer, 2005.
Amills, M., Clop, A., Ramírez, O., and Pérez-Enciso, M., Origin and genetic diversity of pig breeds, Encyclopedia of Life Sciences, Chichester: Wiley, 2010, pp. 1—10. https://doi.org/10.1002/9780470015902.a0022884.
Biscarini, F., Nicolazzi, E.L., Stella, A., et al., Challenges and opportunities in genetic improvement of local livestock breeds, Front. Genet., 2015, vol. 6, no. 33. https://doi.org/10.3389/fgene.2015.00033
Poklukar, K., Čandek-Potokar, M., Batorek Lukač, N., et al., Lipid deposition and metabolism in local and modern pig breeds: a review, Animals, 2020, vol. 10, p. 424. https://doi.org/10.3390/ani10030424
Ernst, L.K., Dmitriev, N.G., and Paronyan, I.A., Genetic Resources of Farm Animals in Russia and Neighboring Countries, Vserossiiskii Nauchno-Issledovatel’skii Institut Genetiki i Razvedeniya Sel’skokhozyaystvennykh Zhivotnykh, 1994.
Koziner, A.B. and Shtakelberg, E.R., Animal Genetic Resources of the USSR, Rome: FAO and UNEP, 1989.
Traspov, A., Deng, W., Kostyunina, O., et al., Population structure and genome characterization of local pig breeds in Russia, Belorussia, Kazakhstan and Ukraine, Genet. Sel. Evol., 2016, vol. 16. https://doi.org/10.1186/s12711-016-0196-y
FAO. http://www.fao.org/dad-is/data/ru/. Accessed November 2, 2021.
Fang, M. and Andersson, L., Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication, Proc. Biol. Sci., 2006, vol. 273, pp. 1803—1810. https://doi.org/10.1098/rspb.2006.3514
Hammond, K. and Leitch, H.W., Genetic resources and the global programme for their management, in The Genetics of the Pig, Rothschild, M.F. and Ruvinsky, A., Eds., Wallingford: CABI, 1998, pp. 405—425.
Woelders, H., Zuidberg, C.A., and Hiemstra, S.J., Animal genetic resources conservation in the Netherlands and Europe: poultry perspective, Poult. Sci., 2005, vol. 85, pp. 216—222. https://doi.org/10.1093/ps/85.2.216
Zhang, J., Jiao, T., and Zhao, S., Genetic diversity in the mitochondrial DNA D-loop region of global swine (Sus scrofa) populations, Biochem. Biophys. Res. Commun., 2016, vol. 473, pp. 814—820. https://doi.org/10.1016/j.bbrc.2016.03.125
Nguyen, H.D., Bui, T.A., Nguyen, P.T., et al., The complete mitochondrial genome sequence of the indigenous I pig (Sus scrofa) in Vietnam, Asian-Australas. J. Anim. Sci., 2017, vol. 30, no. 7, pp. 930—937. https://doi.org/10.5713/ajas.16.0608
Giuffra, E., Kijas, J.M., Amarger, V., et al., The origin of the domestic pig: independent domestication and subsequent introgression, Genetics, 2000, vol. 154, pp. 1785—1791. https://doi.org/10.1093/genetics/154.4.1785
Larson, G., Dobney, K., Albarella, U., et al., Worldwide phylogeography of wild boar reveals multiple centers of pig domestication, Science, 2005, vol. 307, pp. 1618—1621. https://doi.org/10.1126/science.1106927
Wu, G.S., Yao, Y.G., Qu, K.X., et al., Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia, Genome Biol., 2007, vol. 8, no. 11, p. 245. https://doi.org/10.1186/gb-2007-8-11-r245
Ge, Q., Gao, C., Cai, Y., et al., Evaluating genetic diversity and identifying priority conservation for seven Tibetan pig populations in China based on the mtDNA D-loop, Asian-Australas. J. Anim. Sci., 2020, vol. 33, no. 12, pp. 1905—1911. https://doi.org/10.5713/ajas.19.0752
Laval, G., Iannuccelli, N., Legault, C., et al., Genetic diversity of eleven European pig breeds, Genet. Sel. Evol., 2000, vol. 32, no. 2, pp. 187—203. https://doi.org/10.1186/1297-9686-32-2-187
Ji, Y.-Q., Wu, D.-D., Wu, G.-S., et al., Multi-locus analysis reveals a different pattern of genetic diversity for mitochondrial and nuclear DNA between wild and domestic pigs in East Asia, PLoS One, 2011, vol. 6, no. 10, e26416. https://doi.org/10.1371/journal.pone.0026416
Quan, J., Gao, C., Cai, Y., et al., Population genetics assessment model reveals priority protection of genetic resources in native pig breeds in China, Global Ecol. Conserv., 2020, vol. 21. https://doi.org/10.1016/j.gecco.2019.e00829
BLAST. https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed March 1, 2021.
Edgar, R.C., MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 2004, vol. 32, pp. 1792—1797. https://doi.org/10.1093/nar/gkh340
Kumar, S., Stecher, G., and Tamura, K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, no. 7, pp. 1870—1874. https://doi.org/10.1093/molbev/msw054
Castresana, J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis, Mol. Biol. Evol., 2000, vol. 17, pp. 540—552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Peng, M.-S., Fan, L., Shi, N.-N., et al., DomeTree: a canonical toolkit for mitochondrial DNA analyses in domesticated animals, Mol. Ecol. Resour., 2015, vol. 15. https://doi.org/10.1111/1755-0998.12386
Tajima, F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism, Genetics, 1989, vol. 123, pp. 585—595. https://doi.org/10.1093/genetics/123.3.585
Fu, Y.-X., Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection, Genet. Soc. Am., 1997, vol. 147, pp. 915—925. https://doi.org/10.1093/genetics/147.2.915
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J.C., et al., DnaSP 6: DNA sequence polymorphism analysis of large data sets, Mol. Biol. Evol., 2017, vol. 34, pp. 3299—3302. https://doi.org/10.1093/molbev/msx248
Rogers, A.R. and Harpending, H., Population growth makes waves in the distribution of pairwise genetic differences, Mol. Biol. Evol., 1992, vol. 9, pp. 552—569. https://doi.org/10.1093/oxfordjournals.molbev.a040727
Wickham, H., Ggplot2: Elegant Graphics for Data Analysis, New York: Springer-Verlag, 2009.
Excoffier, L. and Lischer, H.E.L., Arlequin Suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows, Mol. Ecol. Resour., 2010, vol. 10, pp. 564—567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Bandelt, H.J., Forster, P., and Röhl, A., Median-joining networks for inferring intraspecific phylogenies, Mol. Biol. Evol., 1999, vol. 16, pp. 37—48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Leigh, N., Georgiev, I., Boeker, T., et al., Nuclear star cluster formation in energy—space, Mon. Not. R. Astron. Soc., 2015, vol. 451, no. 1, pp. 859—869. https://doi.org/10.1093/mnras/stv1012
Lanfear, R., Frandsen, P.B., Wright, A.M., et al., PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses, Mol. Biol. Evol., 2017, vol. 34, pp. 772—773. https://doi.org/10.1093/molbev/msw260
Akaike, H., A new look at statistical model identification, IEEE Trans Auto Control, 1974, vol. 19, pp. 716—723. https://doi.org/10.1109/TAC.1974.1100705
Huson, D. and Kloepper, T., Beyond galled trees—decomposition and computation of galled networks, Eleventh Annual International Conference on Research in Computational Molecular Biology, Oakland, CA, 2007. https://doi.org/10.1007/978-3-540-71681-5_15.
Kharzinova, V.R. and Zinovieva, N.A., The pattern of genetic diversity of different breeds of pigs based on microsatellite analysis, Vavilov J. Genet. Breed., 2020, vol. 24, no. 7, pp. 747—754. https://doi.org/10.18699/VJ20.669
Kharzinova, V.R., Kostyunina, O.V., Karpushkina, T.V., et al., The study of the population structure and genetic diversity of Hungarian Mangalica breed of pigs based on microsatellites analysis, Agrar. Bull. Urale, 2019, vol. 186, no. 7, pp. 77—81. https://doi.org/10.32417/article_5d52b081b3e348.43320197
Balatsky, V.N., Saienko, A.M., Pena, R.N., et al., Genetic diversity of pig breeds on ten production quantitative traits loci, Cytol. Genet., 2015, vol. 49, pp. 299—307. https://doi.org/10.3103/S0095452715050023
Kharzinova, V.R., Kostyunina, O.V., and Zinovieva, N.A., Comparative characterization of the allele pool of local pig breeds based on microsatellite analysis, Pig Breed., 2017, vol. 1, pp. 5—7.
Getmantseva, L., Bakoev, S., Bakoev, N., et al., Mitochondrial DNA diversity in Large White pigs in Russia, Animals, 2020, vol. 10, p. 1365. https://doi.org/10.3390/ani10081365
Wang, C., Chen, Y., Han, J., et al., Mitochondrial DNA diversity and origin of indigenous pigs in South China and their contribution to western modern pig breeds, J. Integr. Agric., 2019, vol. 18, no. 10, pp. 2338—2350. https://doi.org/10.1016/s2095-3119(19)62731-0
Ramírez, O., Ojeda, A., Tomàs, A., et al., Integrating Y-chromosome, mitochondrial, and autosomal data to analyze the origin of pig breeds, Mol. Biol. Evol., 2009, vol. 26, no. 9, pp. 2061—2072. https://doi.org/10.1093/molbev/msp118
Ajibike, A.B., Ilori, B.M., Akinola, O., et al., Assessing the genetic diversity of South-western Nigerian indigenous pig (Sus scorfa) using mitochondrial DNA D-loop sequence, Niger. J. Anim. Sci., 2020, vol. 22, pp. 1—9.
Goodall-Copestake, W., Tarling, G., and Murphy, E., On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals, Heredity, 2012, vol. 109, pp. 50—56. https://doi.org/10.1038/hdy.2012.12
Nei, M. and Tajima, F., DNA polymorphism detectable by restriction endonucleases, Genetics, 1981, vol. 97, p. 145. https://doi.org/10.1093/genetics/97.1.145
Thompson, J.D., Gibson, T.J., Plewniak, F., et al., The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools, Nucleic Acids Res., 1997, vol. 25, pp. 4876—4882. https://doi.org/10.1093/NAR/25.24.4876
Zhang, J., Yang, B., Wen, X., and Sun, G., Genetic variation and relationships in the mitochondrial DNA D-loop region of Qinghai indigenous and commercial pig breeds, Cell Mol. Biol. Lett., 2018, vol. 3, pp. 23—31. https://doi.org/10.1186/s11658-018-0097-x
Tsai, T.S., Rajasekar, S., and John, J.S.St., The relationship between mitochondrial DNA haplotype and the reproductive capacity of domestic pigs (Sus scrofa domesticus), BMC Genet., 2016, vol. 17, р. 67.https://doi.org/10.1186/s12863-016-0375-4
Collingbourne, S.J., Conservation genetics of traditional and commercial pig breeds, and evaluation of their crossbreeding potential for productivity improvement, PhD Thesis, Univ. Essex, 2019.
Moon, K.-H., Nakanishi, M., Futagami, Y., and Kashiwadani, H., Studies on Cambodian species of Graphidaceae (Ostropales, Ascomycota): II, J. Jpn. Bot., 2015, vol. 90, no. 2, pp. 98—102.
Avise, J.C., Phylogeography: The History and Formation of Species, Cambridge, MA, 2000. https://doi.org/10.5860/choice.37-5647.
Kim, T.H., Kim, K.S., and Choi, B.H., Genetic structure of pig breeds from Korea and China using microsatellite loci analysis, J. Anim. Sci., 2005, vol. 83, pp. 2255—2263. https://doi.org/10.2527/2005.83102255X
Kim, K.S., Yeo, J.S., and Kim, J.W., Assessment of genetic diversity of Korean native pig (Sus scrofa) using AFLP markers, Gen. Genet. Syst., 2002, vol. 77, pp. 361—368. https://doi.org/10.1266/GGS.77.361
Li, K.Y., Chen, C., Moran, B.F., et al., Analysis of diversity and genetic relationships between four Chinese indigenous pig breeds and one Australian commercial pig breed, Anim. Genet., 2000, vol. 31, pp. 322—325. https://doi.org/10.1046/j.1365-2052.2000.00649.x
Sundari, S., Chanthran, D., Lim, P.-E., et al., Genetic diversity and population structure of Terapon jarbua (Forskål, 1775) (Teleostei, Terapontidae) in Malaysian waters, ZooKeys, 2020, vol. 911, pp. 139—160. https://doi.org/10.3897/zookeys.911.39222
Stajich, J. and Hahn, M.W., Disentangling the effects of demography and selection in human history, Mol. Biol. Evol., 2004, vol. 22, pp. 63—73. https://doi.org/10.1093/MOLBEV/MSH252
Alexandrino, J., Arntzen, J.W., and Ferrand, N., Nested clade analysis and the genetic evidence for population expansion in the phylogeography of the golden-striped salamander, Chioglossa lusitanica (Amphibia: Urodela), Heredity, 2002, vol. 88, pp. 66—74. https://doi.org/10.1038/sj.hdy.6800010
Li, D., Wei, H., Zhang, Z., et al., Oriental reed warbler (Acrocephalus orientalis) nest defense behaviour to-wards brood parasites and nest predators, Behaviour, 2015. https://doi.org/10.1163/1568539X-00003295
ACKNOWLEDGMENTS
During the study, the equipment of the Center for Collective Use Bioresources and Bioengineering of Agricultural Animals of the Ernst Federal Research Center for Animal Husbandry was used.
Funding
The work was financially supported by the Ministry of Science and Higher Education of the Russian Federation, grant no. 075-15-2021-1037 (internal no. 15.BRC.21.0001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement on the welfare of animals. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Translated by A. Barkhash
Rights and permissions
About this article
Cite this article
Kharzinova, V.R., Akopyan, N.A., Dotsev, A.V. et al. Genetic Diversity and Phylogenetic Relationships of Russian Pig Breeds Based on the Analysis of mtDNA D-Loop Polymorphism. Russ J Genet 58, 944–954 (2022). https://doi.org/10.1134/S102279542208004X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102279542208004X