Abstract
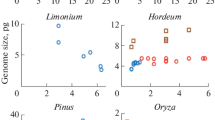
Genome size in plant species is closely associated with many intracellular and environmental factors, which makes it important for adaptation and evolution. Although research on genome size changes has a long history, the problem continues to be challenging because many factors affecting this evolutionary process are still unknown. Despite substantial effort to investigate genome size changes in major plant lineages, the present-day picture remains rather static because the age of plant species is not always taken into consideration. We attempted to systematically investigate genome size dynamics in multiple genera of diploid seed plants. A remarkably strong intrageneric linear dependence between the genome size and evolutionary age of species was found. This linearity of intrageneric genome size dynamics has persisted over millions of years, suggesting gradual changes in DNA content unrelated to the natural selection process. Our results also reveal that some outlier species did not follow this general trend and could go through drastic changes in genome size over a short period of evolutionary time. We conclude that genome size within the genera of many diploid seed plant species is not stochastic and follows a linear dependence on the evolutionary age of the species.


Similar content being viewed by others
REFERENCES
Bennett, M.D. and Leitch, I.J., Plant genome size research: a field in focus, Ann. Bot., 2005, vol. 95, pp. 1—6.
Greilhuber, J., Dolezel, J., Lysák, M.A., and Bennett, M.D., The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents, Ann. Bot., 2005, vol. 95, pp. 255—260.
Petrov, D.A., Mutational equilibrium model of genome size evolution, Theor. Popul. Biol., 2002, vol. 61, pp. 531—544.
Muotri, A.R., Marchetto, M.C., Coufal, N.G., and Gage, F.H., The necessary junk: new functions for transposable elements, Hum. Mol. Genet., 2007, vol. 16, no. 2, pp. R159—R167.
Jurka, J., Kapitonov, V.V., Kohany, O., and Jurka, M.V., Repetitive sequences in complex genomes: structure and evolution, Annu. Rev. Genomics Hum. Genet., 2007, vol. 8, pp. 241—259.
Jurka, J., Conserved eukaryotic transposable elements and the evolution of gene regulation, Cell Mol. Life Sci., 2008, vol. 65, no. 2, pp. 201—204.
Palazzo, A.F. and Gregory, T.R., The case for junk DNA, PLoS Genet., 2014, vol. 10. e1004351. https://doi.org/10.1371/journal.pgen.1004351
Dodsworth, S., Leitch, A.R., and Leitch, I.J., Genome size diversity in angiosperms and its influence on gene space, Curr. Opin. Genet. Dev., 2015, vol. 35, pp. 73—78. https://doi.org/10.1016/j.gde.2015.10.006
Pellicer, J., Hidalgo, O., Dodsworth, S., and Leitch, I.J., Genome size diversity and its impact on the evolution of land plant, Genes (Basel), 2018, E88. https://doi.org/10.3390/genes9020088
Cavalier-Smith, T., Skeletal DNA and the evolution of genome size, Annu. Rev. Biophys. Bioeng., 1982, vol. 11, pp. 273—302.
Cavalier-Smith, T., Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion, Ann. Bot., 2005, vol. 95, pp. 147—175.
Gregory, T.R., Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma, Biol. Rev., 2001, vol. 7, pp. 65—101.
Vinogradov, A.E., Genome size and chromatin condensation in vertebrates, Chromosoma, 2005, vol. 113, pp. 362—369.
Alonso, C., Pérez, R., Bazaga, P., and Herrera, C.M., Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms, Front. Genet., 2015, vol. 6, no. 4. https://doi.org/10.3389/fgene.2015.00004
Morgan, H.D. and Westoby, M., The relationship between nuclear DNA content and leaf strategy in seed plants, Ann. Bot., 2005, vol. 96, pp. 1321—1330.
Garnatje, T., Vallès, J., Garcia, S., et al., Genome size in Echinops L. and related genera (Asteraceae, Cardueae): karyological, ecological and phylogenetic implications, Biol. Cell, 2004, vol. 96, pp. 117—124.
Gregory, T.R., Genome size and developmental complexity, Genetica, 2002, vol. 115, no. 1, pp. 131—146.
Markov, A.V., Anisimov, V.A., and Korotayev, A.V., Relationship between genome size and organismal complexity in the lineage leading from prokaryotes to mammals, Paleontol. J., 2010, vol. 44, no. 5, pp. 363—373. https://doi.org/10.1134/S0031030110040015
Vinogradov, A.E. and Anatskaya, O.V., Genome size and metabolic intensity in tetrapods: a tale of two lines, Proc. Biol. Sci., 2006, vol. 273, pp. 27—32.
Chénais, B., Caruso, A., Hiard, S., and Casse, N., The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments, Gene, 2012, vol. 509, pp. 7—15. https://doi.org/10.1016/j.gene.2012.07.042
Leitch, A.R. and Leitch, I.J., Ecological and genetic factors linked to contrasting genome dynamics in seed plants, New Phytol., 2012, vol. 194, pp. 629—646. https://doi.org/10.1111/j.1469-8137.2012.04105.x
Knight, C.A., Molinari, N.A., and Petrov, D.A., The large genome constraint hypothesis: evolution, ecology and phenotype, Ann. Bot., 2005, vol. 95, pp. 177—190.
Hidalgo, O., Pellicer, J., Christenhusz, M., et al., Is there an upper limit to genome size? Trends Plant Sci., 2017, vol. 22, no. 7, pp. 567—573. https://doi.org/10.1016/j.tplants.2017.04.005
Guignard, M.S., Crawley, M.J., Kovalenko, D., et al., Interactions between plant genome size, nutrients and herbivory by rabbits, molluscs and insects on a temperate grassland, Proc. Biol. Sci., 2019, vol. 286, no. 1899, p. 20182619. https://doi.org/10.1098/rspb.2018.2619
Sheremetiev, S.N., Gamalei, Yu.V., and Slemnev, N.N., Trends of angiosperm genome evolution, Tsitologiya, 2011 vol. 53, no. 4, pp. 295—312.
Vinogradov, A.E., Selfish DNA is maladaptive: evidence from the plant Red list, Trends Genet., 2003, vol. 19, pp. 609—614.
Petrov, D.A., Evolution of genome size: new approaches to an old problem, Trends Genet. 2001, vol. 17, pp. 23—28.
Vitte, C. and Panaud, O., LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model, Cytogenet. Genome Res., 2005, vol. 110, pp. 91—107.
Ågren, J.A. and Wright, S.I., Co-evolution between transposable elements and their hosts: a major factor in genome size evolution?, Chromosome Res., 2011, vol. 19, pp. 777—786. https://doi.org/10.1007/s10577-011-9229-0
Zhao, M. and Ma, J., Co-evolution of plant LTR-retrotransposons and their host genomes, Protein Cell, 2013, vol. 4, pp. 493—501. https://doi.org/10.1007/s13238-013-3037-6
Elliott, T.A. and Gregory, T.R., Do larger genomes contain more diverse transposable elements?, BMC Evol. Biol., 2015, vol. 5, p. 69. https://doi.org/10.1186/s12862-015-0339-8
Devos, K.M., Brown, J.K., and Bennetzen, J.L., Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis,Genome Res., 2002, vol. 12, pp. 1075—1079.
Bennetzen, J.L., Mechanisms and rates of genome expansion and contraction in flowering plants, Genetica, 2002, vol. 115, pp. 29—36.
Orel, N. and Puchta, H., Differences in the processing of DNA ends in Arabidopsis thaliana and tobacco: possible implications for genome evolution, Plant Mol. Biol., 2003, vol. 51, pp. 523—531.
Gregory, T.R., Is small indel bias a determinant of genome size?, Trends Genet., 2003, vol. 19, no. 9, pp. 485—488.
Puchta, H., The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution, J. Exp. Bot., 2005, vol. 56, pp. 1—14.
Leitch, I.J., Chase, M.W., and Bennett, M.D., Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants, Ann. Bot., 1998, vol. 82, pp. 85—94.
Soltis, D.E., Soltis, P.S., Bennett, M.D., and Leitch, I.J., Evolution of genome size in the angiosperms, Am. J. Bot., 2003, vol. 90, pp. 1596—1603.
Leitch, I.J., Soltis, D.E., Soltis, P.S., and Bennett, M.D., Evolution of DNA amounts across land plants (Embryophyta), Ann. Bot., 2005, vol. 95, pp. 207—217.
Leitch, I.J., Beaulieu, J.M., Chase, M.W., et al., Genome size dynamics and evolution in monocots, J. Bot., 2010, article ID 862516. https://doi.org/10.1155/2010/862516
Wendel, J.F., Cronn, R.C., Johnston, J.S., and Price, H.J., Feast and famine in plant genomes, Genetica, 2002, vol. 115, pp. 37—47.
Johnston, J.S., Pepper, A.E., Hall, A.E., Chen, Z.J., Hodnett, G., Drabek, J., et al., Evolution of genome size in Brassicaceae, Ann. Bot., 2005, vol. 95, pp. 229—235.
Price, H.J., Dillo, S.L., Hodnett, G., et al., Genome evolution in the genus Sorghum (Poaceae), Ann. Bot., 2005, vol. 95, pp. 219—227.
Leitch, I.J., Beaulieu, J.M., Cheung, K., et al., Punctuated genome size evolution in Liliaceae, J. Evol. Biol., 2007, vol. 20, pp. 2296—2308.
Lysak, M.A., Koch, M.A., Beaulieu, J.M., et al., The dynamic ups and downs of genome size evolution in Brassicaceae, Mol. Biol. Evol., 2009, vol. 26, pp. 85—98. https://doi.org/10.1093/molbev/msn223
Leitch, J., Kahandawala, I., Suda, J., et al., Genome size diversity in orchids: consequences and evolution, Ann. Bot., 2009, vol. 104, pp. 469—481.https://doi.org/10.1093/aob/mcp003
Pellicer, J., Kelly, L.J., Magdalena, C., and Leitch, I.J., Insights into the dynamics of genome size and chromosome evolution in the early diverging angiosperm lineage Nymphaeales (water lilies), Genome, 2013, vol. 56, pp. 437—449. https://doi.org/10.1139/gen-2013-0039
Pellicer, J., Kelly, L.J., Leitch, I.J., et al., A universe of dwarfs and giants: genome size and chromosome evolution in the monocot family Melanthiaceae, New Phytol., 2014, vol. 201, pp. 1484—1497. https://doi.org/10.1111/nph.12617
Garcia, S., Leitch, I.J., Anadon-Rosell, A., et al., Recent updates and developments to plant genome size databases, Nucleic Acids Res., 2014, vol. 42, pp. D1159—D1166. https://doi.org/10.1093/nar/gkt1195
Berry, P.E., Hahn, W.J., Sytsma, K.J., et al., Phylogenetic relationships and biogeography of Fuchsia (Onagraceae) based on noncoding nuclear and chloroplast DNA data, Am. J. Bot., 2004, vol. 91, pp. 601—614.
Lledó, M.D., Molecular phylogenetics of Limonium and related genera (Plumbaginaceae): biogeographical and systematic implications, Am. J. Bot. 2003, vol. 92, pp. 1189—1198.
Good-Avila, S.V., Timing and rate of speciation in Agave (Agavaceae), Proc. Natl. Acad. Sci. U.S.A., 2006, vol. 103, pp. 9124—9129.
Moore, B.R. and Donoghue, M.J., Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations, Am. Nat., 2007, vol. 170, suppl. 2, pp. S28—S55. https://doi.org/10.1086/519460
Qiao, C.-Y., Ran, J.H., Li, Y., and Wang, X.Q., Phylogeny and biogeography of Cedrus (Pinaceae) inferred from sequences of seven paternal chloroplast and maternal mitochondrial DNA regions, Ann. Bot., 2007, vol. 100, pp. 573—580.
Guzmán, B., Lledó, M.D., and Vargas, P., Adaptive radiation in Mediterranean Cistus (Cistaceae), PLoS One, 2009, vol. 4. e6362. https://doi.org/10.1371/journal.pone.0006362
Blattner, F.R., Pleines, T., and Jakob, S.S., Rapid radiation in the barley genus Hordeum (Poaceae) during the Pleistocene in the Americas, in Evolution in Action, 2010. https://doi.org/10.1007/978-3-642-12425-9_2
Carlsen, M.M., Understanding the origin and rapid diversification of the genus Anthurium Schott (Araceae), integrating molecular phylogenetics, morphology and fossils, PhD Thesis, Graduate School at the University of Missouri, USA. 2011. ISBN: 9781124890043.
Poczai, P., Molecular genetic studies on complex evolutionary processes in Archaesolanum (Solanum, Solanaceae), PhD Thesis,University of Pannonia,Hungary, 2011. http://konyvtar.uni-pannon.hu/doktori/2011/ Poczai_Peter_dissertation.pdf.
Sanz, M., Schneeweiss, G.M., Vilatersana, R., and Vallès, J., Temporal origins and diversification of Artemisia and allies (Anthemideae, Asteraceae), Collect. Bot., 2011, vol. 30, pp. 7—15.
Ikinci, N., Molecular phylogeny and divergence times estimates of Lilium section Liriotypus (Liliaceae) based on plastid and nuclear ribosomal ITS DNA sequence data. Turk. J. Bot., 2011, vol. 35, pp. 319—330. https://doi.org/10.3906/bot-1003-29
Guo, Y.Y., Luo, Y.B., Liu, Z.J., and Wang, X.Q., Evolution and biogeography of the slipper orchids: Eocene vicariance of the conduplicate genera in the Old and New World tropics, PLoS One, 2012, vol. 7, no. 6. e38788. https://doi.org/10.1371/journal.pone.0038788
Lo, E.Y.Y. and Donoghue, M.J., Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae), Mol. Phylogenet. Evol., 2012, vol. 63, pp. 230—243. https://doi.org/10.1016/j.ympev.2011.10.005
Heibl, C. and Renner, S.S., Distribution models and a dated phylogeny for Chilean Oxalis species reveal occupation of new habitats by different lineages, not rapid adaptive radiation, Syst. Biol., 2012, vol. 61, no. 5, pp. 823—834. https://doi.org/10.1093/sysbio/sys034
Bliss, B.J. and Suzuki, J.Y., Genome size in Anthurium evaluated in the context of karyotypes and phenotypes, AoB Plants, 2012, pls006. https://doi.org/10.1093/aobpla/pls006
Díez, C.M., Gaut, B.S., Meca, E., Scheinvar, E., Montes-Hernandez, S., Eguiarte, L.E., et al., Genome size variation in wild and cultivated maize along altitudinal gradients, New Phytol., 2013, vol. 199, pp. 264—276. https://doi.org/10.1111/nph.12247
Wan, Y., Schwaninger, H.R., Baldo, A.M., Labate, J.A., Zhong, G.-Y., and Simon, C.J., A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change, BMC Evol. Biol., 2013, vol. 13, no. 141. https://doi.org/10.1186/1471-2148-13-141
McLeish, M.J., Miller, J.T., and Mound, L.A., Delayed colonization of Acacia by thrips and the timing of host-conservatism and behavioural specialization, BMC Evol. Biol., 2013, vol. 13, no. 188. https://doi.org/10.1186/1471-2148-13-188
Lockwood, J.D., Aleksić, J.M., Zou, J., Wang, J., Liu, J., and Renner, S.S., A new phylogeny for the genus Picea from plastid, mitochondrial, and nuclear sequences, Mol. Phylogenet. Evol., 2013 vol. 69, pp. 717—727. https://doi.org/10.1016/j.ympev.2013.07.004
Wang, B., Hybridization and evolution in the genus Pinus, PhD Thesis, Umeå University, Sweden, 2013. https://www.diva-portal.org/smash/get/diva2:652236/ FULLTEXT01.pdf.
Sherman-Broyles, S., Bombarely, A., Grimwood, J., et al., Complete plastome sequences from Glycine syndetika and Six additional perennial wild relatives of soybean,G3 (Bethesda), 2014, vol. 4, pp. 2023—2233. https://doi.org/10.1534/g3.114.012690
Kranitz, M.L., Biffin, E., Clark, A., et al., Evolutionary diversification of New Caledonian Araucaria,PLoS One, 2014, vol. 29, no. 10. e110308. https://doi.org/10.1371/journal.pone.0110308
Fougère-Danezan, M., Joly, S., Bruneau, A., et al., Phylogeny and biogeography of wild roses with specific attention to polyploids, Ann. Bot., 2015, vol. 115, pp. 275—291. https://doi.org/10.1093/aob/mcu245
Zonneveld, B.J., Leitch, I.J., and Bennett, M.D., First nuclear DNA amounts in more than 300 angiosperms, Ann. Bot., 2005, vol. 96, no. 2, pp. 229—244.
Bai, C., Alverson, W.S., Follansbee, A., and Waller, D.M., New reports of nuclear DNA content for 407 vascular plant taxa from the United States, Ann Bot., 2002, vol. 110, no. 8, pp. 1623—1629. https://doi.org/10.1093/aob/mcs222
Graham, M.J., Nickell, C.D., and Rayburn, A.L., Relationship between genome size and maturity group in soybean, Theor. Appl. Genet., 1994, vol. 88, pp. 429—432.
Realini, M.F., Poggio, L., Cámara-Hernández, J., and González, G.E., Intra-specific variation in genome size in maize: cytological and phenotypic correlates, AoB Plants, 2015, vol. 8, plv138. https://doi.org/10.1093/aobpla/plv138
Díez, C.M., Gaut, B.S., Meca, E., et al., Genome size variation in wild and cultivated maize along altitudinal gradients, New Phytol., 2013, vol. 199, no. 1, pp. 264—276. https://doi.org/10.1111/nph.12247
Garcia, S., Garnatje, T., Twibell, J.D., and Vallès, J., Genome size variation in the Artemisia arborescens complex (Asteraceae, Anthemideae) and its cultivars, Genome, 2006, vol. 49, pp. 244—253.
Gunn, B.F., Baudouin, L., Beulé, T., et al., Ploidy and domestication are associated with genome size variation in palms, Am. J. Bot., 2015, vol. 102, pp. 1625—1633. https://doi.org/10.3732/ajb.1500164
Greilhuber, J. and Ebert, I., Genome size variation in Pisum sativum,Genome, 1994, vol. 37, no. 4, pp. 646—655.
Piegu, B., Guyot, R., Picault, N., et al., Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice, Genome Res., 2006, vol. 16, pp. 1262—1269.
Bennetzen, J.L., Ma, J., and Devos, K.M., Mechanisms of recent genome size variation in flowering plants, Ann Bot., 2005, vol. 95, pp. 127—132.
Vinogradov, A.E., Evolution of genome size: multilevel selection, mutation bias or dynamical chaos?, Curr. Opin. Genet. Dev., 2006, vol. 14, pp. 620—626.
Funding
This work was supported by funds from United States Department of Agriculture, Agricultural Research Service, Beltsville, Maryland (CRIS project 8042-21000-268-00D).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Supplementary material
Rights and permissions
About this article
Cite this article
Boutanaev, A.M., Nemchinov, L.G. Genome Size Dynamics within Multiple Genera of Diploid Seed Plants. Russ J Genet 56, 684–692 (2020). https://doi.org/10.1134/S1022795420060046
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420060046