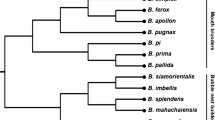
Abstract
Sheep were one of the first animals to be domesticated. The history of sheep domestication and their widespread distribution dates to about ten thousand years ago, during which sheep exhibit both physical changes and modifications at the genetic level. The authors developed a system of 49 oligonucleotide primers for targeted Next Generation Sequencing (NGS) of genetic loci for phylogenetic analysis and identifying economically useful traits. Altogether, NGS libraries were prepared and sequenced on an Illumina MiSeq platform(Illumina) for 48 samples, for 40 of which it was possible to determine phylogenetic lineages: 28 belonged to haplogroup B, 10 to haplogroup A, and one sample each to haplogroups C and D. Study of the genes associated with economically useful traits revealed the samples with nucleotide substitutions in the MC1R gene leading to black coat color: two samples with c.218T>A, one with c.361G>A, and two with both substitutions simultaneously, as well as one sample with the substitution in the GDF8 gene associated with muscle hypertrophy and one with the substitution in the TYRP1 gene associated with brown coat color. The data obtained confirm a high genetic diversity of sheep from ancient southwestern Siberia and the utility of targeted sequencing for the study of ancient DNA samples.





Similar content being viewed by others
REFERENCES
Vigne, J.D., The origins of animal domestication and husbandry: a major change in the history of humanity and the biosphere, C. R. Biol., 2011, vol. 334, no. 3, pp. 171—181. https://doi.org/10.1016/j.crvi.2010.12.009
Zeder, M.A., Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact, Proc. Natl. Acad. Sci. U.S.A., 2008, vol. 105, no. 33, pp. 11597—11604. https://doi.org/10.1073/pnas.0801317105
Ryder, M., Sheep and Man, Duckworth, 1983.
Meadows, J.R.S., Hiendleder, S., and Kijas, J.W., Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel, Heredity (Edinburgh), 2011, vol. 106, no. 4, pp. 700—706. https://doi.org/10.1038/hdy.2010.122
Ermolenko, N.A., Boyarskikh, U.A., Kechin, A.A., et al., Massive parallel sequencing for diagnostic genetic testing of BRCA genes—a single center experience, Asian Pac. J. Cancer Prev., 2015, vol. 16, no. 17, pp. 7935—7941. https://doi.org/10.7314/apjcp.2015.16.17.7935
Pääbo, S., Gifford, J.A., and Wilson, A.C., Mitochondrial DNA sequences from a 7000-year old brain, Nucleic Acids Res., 1988, vol. 16, no. 20, p. 9775. https://doi.org/10.1093/nar/16.20.9775
Nguyen-Dumont, T., Pope, B.J., Hammet, F., et al., A high-plex PCR approach for massively parallel sequencing, Biotechniques, 2013, vol. 55, no. 2, pp. 69—74. https://doi.org/10.2144/000114052
Bolger, A.M., Lohse, M., and Usadel, B., Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, vol. 30, no. 15, pp. 2114—2120. https://doi.org/10.1093/bioinformatics/btu170
Li, H. and Durbin, R., Fast and accurate short read alignment with Burrows—Wheeler transform, Bioinformatics, 2009, vol. 25, no. 14, pp. 1754—1760. https://doi.org/10.1093/bioinformatics/btp324
Kechin, A., Boyarskikh, U., Kel, A., and Filipenko, M., cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing, J. Comput. Biol., 2017, vol. 24, no. 11, pp. 1138—1143. https://doi.org/10.1089/cmb.2017.0096
Dymova, M.A., Zadorozhny, A.V., Mishukova, O.V., et al., Mitochondrial DNA analysis of ancient sheep from Altai, Anim. Genet., 2017, vol. 48, no. 5, pp. 615—618. https://doi.org/10.1111/age.12569
Tamura, K., Stecher, G., Peterson, D., et al., MEGA6: molecular evolutionary genetics analysis version 6.0, Mol. Biol. Evol., 2013, vol. 30, no. 12, pp. 2725—2729. https://doi.org/10.1093/molbev/mst197
Clop, A., Marcq, F., Takeda, H., et al., A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep, Nat. Genet., 2006, vol. 38, no. 7, pp. 813—818. https://doi.org/10.1038/ng1810
Hinten, G.N., Hale, M.C., Gratten, J., et al., SNP-SCALE: SNP scoring by colour and length exclusion, Mol. Ecol. Notes, 2007, vol. 7, no. 3, pp. 377—388. https://doi.org/10.1111/j.1471-8286.2006.01648.x
Stiller, M., Knapp, M., Stenzel, U., et al., Direct multiplex sequencing (DMPS)—a novel method for targeted high-throughput sequencing of ancient and highly degraded DNA, Genome Res., 2009, vol. 19, no. 10, pp. 1843—1848. https://doi.org/10.1101/gr.095760.109
Kechin, A., Khrapov, E., Boyarskikh, U., et al., BRCA-analyzer: automatic workflow for processing NGS reads of BRCA1 and BRCA2 genes, Comput. Biol. Chem., 2018, vol. 77, pp. 297—306. https://doi.org/10.1016/j.compbiolchem.2018.10.012
Tapio, M., Marzanov, N., Ozerov, M., et al., Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas, Mol. Biol. Evol., 2006, vol. 23, no. 9, pp. 1776—1783. https://doi.org/10.1093/molbev/msl043
Lv, F.-H., Peng, W.-F., Yang, J., et al., Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep, Mol. Biol. Evol., 2015, vol. 32, no. 10, pp. 2515—2533. https://doi.org/10.1093/molbev/msv139
ACKNOWLEDGMENTS
We are grateful to D.V. Panin and Ya.V. Frolov for the samples from archaeological sites Rublevo-VI and Firsovo-XIV provided for the comparative analysis and to the native English speaker T.R. Hermes for thorough text revision.
Funding
This work was supported by the Russian Science Foundation (project no. 16-18-10033 “Formation and Evolution of Life Support Systems in Nomadic Societies of Altai and Adjacent Territories in the Late Antiquity and Middle Ages: Complex Reconstruction”).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by A. Barkhash
Supplementary material
Rights and permissions
About this article
Cite this article
Kechin, A.A., Dymova, M.A., Tishkin, A.A. et al. Targeted Sequencing for Studying Economically Useful Traits and Phylogenetic Diversity of Ancient Sheep. Russ J Genet 55, 1499–1505 (2019). https://doi.org/10.1134/S102279541912007X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102279541912007X