Abstract
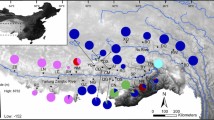
Population structuring of a species provide basic information for biological conservation. We investigated the genetic structure of seven populations of Rana pseudodalmatina, an endemic species of the Ranidae inhabiting the Hyrcanian forests in northern Iran, based on the genetic variation of two partial mitochondrial DNA sequences (16S rRNA and cytochrome b genes). Molecular genetic analyses revealed remarkable variation among populations of R. pseudodalmatina. The phylogenetic trees clearly indicated two distinct haplogroups, which largely corresponded to their geographic locations. A strong population structure was found (ΦCT = 0.559, P = 0.027) with high haplotype diversity (Hd = 0.88) and low nucleotide diversity (π = 0.0041). Analysis of molecular variance (AMOVA) indicated that most of the observed genetic variation (55.92%) occurred between the two haplogroups. Also, Mantel tests revealed a significant correlation between geographic and genetic distances (R2 = 0.33, P = 0.005). Finally, the star-like topology of haplotype network and also neutrality tests provide strong evidence for population expansion in two haplogroups. All the findings of the present study, suggest a strong evidence for past expansion of isolated populations of R. pseudodalmatina, which their isolation could be largely attributed to rising level of the Caspian Sea during last glacial periods in the Pleistocene.



Similar content being viewed by others
REFERENCES
Frankham, R., Ballou, J.D. and Briscoe, D.A., Introduction to Conservation Genetics, New York: Cambridge University Press, 2002.
Hughes, A.R., Inouye, B.D., Johnson, M.T.J., et al., Ecological consequences of genetic diversity, Ecol. Lett., 2008, vol. 11, pp. 609-623.
Scherer, R.D., Muths, E., Noon, B.R. and Oyler-McCance, S.J., The genetic structure of a relict population of wood frogs, Conserv. Genet., 2012, vol. 13, pp. 1521-1530.
Newman, D. and Pilson, D., Increased probability of extinction due to decreased genetic effective population size: experimental populations of Clarkia pulchella, Evolution, 1997, vol. 51, pp. 354-362.
Stuart, S.N., Chanson, J.S., Cox, N.A., et al., Status and trends of amphibian declines and extinctions worldwide, Science, 2004, vol. 306, pp. 1783-1786.
Wake, D.B., Facing extinction in real time, Science, 2012, vol. 335, pp.1052-1053.
Cushman, S.A., Effects of habitat loss and fragmentation on amphibians: a review and prospectus, Biol. Conserv., 2006, no. 128, pp. 231-240.
Storfer, A., Eastman, J.M. and Spear, S.F., Modern methods for amphibian conservation, BioScience, 2009, vol. 59, no. 7, pp. 559-571.
Allentof, M.E. and O’Brien, J., Global amphibian declines, loss of genetic diversity and fitness: a review, Diversity, 2010, vol. 2, pp. 47-71.
Nori, J., Lemes, P., Urbina-Cardona, N., et al., Amphibian conservation, land use changes and protected areas: a global overview, Biol. Conserv., 2015, vol. 191, pp. 367-374.
Veith, M., Kosuch, J. and Vences, M., Climatic oscillations triggered post-Messinian speciation of Western Palearctic brown frogs (Amphibia, Anura, Ranidae), Mol. Phylogenet. Evol., 2003, vol. 26, pp. 310-327.
Picariello, O., Feliciello, I., Scillitani, G., et al., Evidenze morfologiche e molecolari dell’identita tassonomica di Rana macrocnemis, Rana camerani e Rana holtzi (Anura: Ranidae), Riv. Idrobiol. (Perugia),1999, vol. 38, pp. 168-182.
Pesarakloo, A., Gharzi, A. and Kami, H.G., An investigation on the reproductive biology of Rana macrocnemis pseudodalmatina (Amphibians: Anura) in Golestan province (Minudasht), Iri. J. Biol., 2009, vol. 25, no. 1, pp. 55-63 [In Farsi].
Bruford, M.W., Hanotte, O., Brookfeild, J.F.Y., et al., Multilocus and single locus DNA fingerprinting, Molecular Genetic Analysis of Populations: A Practical Approach, Hoelzel, A.R., Ed., Oxford, UK: IRL Press, 1998, 2nd ed.
Hall, T.A., BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows95/98/NT, Nucleic Acids Symp. Ser., 1999, vol. 41, pp. 95-98.
Farris, J.S, Källersjö, M., Kluge, A.G., et al., Constructing a significance test for incongruence, Syst. Biol., 1995, vol. 44, pp. 570-572.
Swofford, D.L., PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods): Version 4, Sunderland: Sinauer Associates, 2003.
Librado, P. and Rozas, J., DnaSP v5: a software for comprehensive analysis of DNA polymorphism data, Bioinformatics, 2009, vol. 25, pp. 1451-1452.
Excoffier, L. and Lischer, H.E.L., Arlequin Suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows, Mol. Ecol. Resour., 2010, vol. 10, pp. 564-567. doi 10.1111/j.1755-0998.2010.02847.x
Tajima, F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism, Genetics, 1989, vol. 123, pp. 585-595.
Fu, Y-X., Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection, Genetics,1997, vol. 147, pp. 915-925.
Rousset, F., Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance, Genetics, 1997, vol. 145, pp. 1219-1228.
Hijmans, R. J., Cruz, M., Rojas, E. and Guarino, L., DIVA-GIS, version 1.4: a geographic information system for the management and analysis of genetic resources data, 2001. http://www.diva-gis.org.
Guindon, S. and Gascuel, O., A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood, Syst. Biol., 2003, vol. 52, pp. 696-704.
Posada, D., jModelTest: phylogenetic model averaging, Mol. Biol. Evol., 2008, vol. 25, pp. 1253-1256.
Gascuel, O., BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data, Mol. Biol. Evol., 1997, vol. 14, pp. 685-695.
Ronquist, F. and Huelsenbeck, J.P., MrBayes 3: Bayesian phylogenetic inference under mixed models, Bioinformatics, 2003, vol. 19, pp. 1572-1574.
Ronquist, F., Teslenko, M., Van der Mark, P., et al., MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space, Syst. Biol., 2012, vol. 61, no. 3, pp. 539-542.
Bandelt, H.J., Forster, P. and Röhl, A., Median-joining networks for inferring intraspecific phylogenies, Mol. Biol. Evol., 1999, vol. 16, pp. 37-48.
Farjallah, S., Amor, N., Merella, P., et al., Pattern of genetic diversity of North African green frog Pelophylax saharicus (Amphibia) in Tunisia, Pak. J. Zool., 2012, vol. 44, no. 4, pp. 901-907.
Avise, J.C., Phylogeography: The History and Formation of Species, Cambridge, Massachusetts: Harvard University Press, 2000.
Miller, A.G., Hyrcanian forests, Iran and Azerbaijan, Centres of Plant Diversity: A Guide and Strategy for Their Conservation in 3 Volumes, Davis, S.D., Heywood, V.H., and Hamilton, A.C., Eds., Cambridge, UK: WWF, 1994, vol. 1, pp. 343-344.
Akhani, H., Djamali, M., Ghorbanalizadeh, A., et al., Plant biodiversity of Hyrcanian relict forests, Iran: an overview of the flora, vegetation, palaeoecology and conservation, Pak. J. Bot., 2010, vol. 42, pp. 231–258.
Yanko-Hombach, V., Kroonenberg, S., and Leroy, S.A.G., Caspian-Black Sea-Mediterranean corridors during the last 30 ka: sea level change and human adaptive strategies, Proceedings of IGCP 521 and 481 INQUA 501 Third Plenary Meeting and Field Trip, Quat. Int., 2010, vol. 225, no. 2, pp. 147-149.
Slatkin, M. and Hudsont, R.R., Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations, Genetics, 1991, vol. 129, pp. 555-562.
ACKNOWLEDGMENTS
We would like to thank the Iranian Department of Environment for sampling authorization and support during fieldwork especially Golestan, Mazanderan and Gilan Department of Environment. We wish to thank Razi University authorities for support during fieldwork and collecting specimens. We also thank the honorable authorities of Hakim Sabsevari University for helping with the lab task. We specially thank Seyyed Saeed Hosseinian Yousefkhani for his help in the molecular laboratory and Mr. Amirreza Zarei and Reza Motamedi for their unforgettable help during field work.
Author information
Authors and Affiliations
Corresponding author
Additional information
1The article is published in the original.
Rights and permissions
About this article
Cite this article
Najibzadeh, M., Gharzi, A., Rastegar-Pouyani, N. et al. Genetic Structure of the Hyrcanian Wood Frog, Rana pseudodalmatina (Amphibia: Ranidae) Using mtDNA Gene Sequences. Russ J Genet 54, 1221–1228 (2018). https://doi.org/10.1134/S1022795418100095
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795418100095