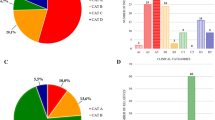
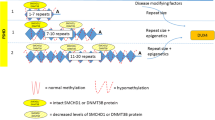
Abstract
Landouzy–Dejerine facioscapulohumeral muscular dystrophy (FSHD) is one of the most common hereditary myodystrophies. A study of the genetic nature of the disease, which has an autosomal dominant mode of inheritance, is extremely interesting and revealing. A unique structure of D4Z4 macrosatellite repeats found in the 4q35 region was originally characterized by a decrease in the number of repeats in patients with Landouzy–Dejerine muscular dystrophy, which resulted in the activation of neighboring genes, in particular, the DUX4 transcription factor. Later, it was found that the epigenetic mechanisms responsible for the chromatin condensation of this region underlie the activation. To date, additional participants leading to pathogenesis of the disease, such as SMCHD1 methylation regulator and DBE-T regulatory long noncoding RNA, have been identified. The revealed complexity of the disease mechanisms is in good agreement with the observed pattern of the disease inheritance. The study of the Landouzy–Dejerine muscular dystrophy pathogenesis is a good example of how monogenic diseases can possess a more complex nature of inheritance.
Similar content being viewed by others
References
Padberg, G.W., Frants, R.R., Brouwer, O.F., et al., Facioscapulohumeral muscular dystrophy in the Dutch population, Muscle Nerve Suppl., 1995, vol. 2, pp. S81–S84.
Flanigan, K.M., Coffeen, C.M., Sexton, L., et al., Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy, Neuromuscular Disord., 2001, vol. 11, nos. 6–7, pp. 525–529.
Mostacciuolo, M.L., Pastorello, E., Vazza, G., et al., Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample, Clin. Genet., 2009, vol. 75, no. 6, pp. 550–555. doi 10.1111/j.1399-0004.2009.01158.x
Norwood, F.L., Harling, C., Chinnery, P.F., et al., Prevalence of genetic muscle disease in Northern England: in-depth analysis of a muscle clinic population, Brain, 2009, vol. 132, no. 11, pp. 3175–3186. doi 10.1093/brain/awp236
Zatz, M., Marie, S.K., Passos-Bueno, M.R., et al., High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families, Am. J. Hum. Genet., 1995, vol. 56, no. 1, pp. 99–105.
Bakker, E., Wijmenga, C., Vossen, R.H., et al., The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter, Muscle Nerve Suppl., 1995, vol. 2, pp. S39–S44.
Landouzy, L. and Dejerine, J., De la myopathie atrophique progressive; myopathie héréditaire, sans neuropathie, debutant d’ordinaire dans l’enfance par la face, Paris: F. Alcan, 1885.
Padberg, G., Facioscapulohumeral Disease, Leiden: Univ. Leiden, 1982.
Tawil, R., Storvick, D., Feasby, T.E., et al., Extreme variability of expression in monozygotic twins with FSH muscular dystrophy, Neurology, 1993, vol. 43, no. 2, pp. 345–348.
Padberg, G.W., Facioscapulohumeral muscular dystrophy: a clinician’s experience, in Facioscapulohumeral Muscular Dystrophy (FSHD): Clinical Medicine and Molecular Cell Biology, London: Garland Science, 2004, pp. 41–51.
Tawil, R., Facioscapulohumeral muscular dystrophy, Neurotherapeutics, 2008, vol. 5, no. 4, pp. 601–606. doi 10.1016/j.nurt.2008.07.005
Quarantelli, M., Lanzillo, R., Del Vecchio, W., et al., Modifications of brain tissue volumes in facioscapulohumeral dystrophy, Neuroimage, 2006, vol. 32, no. 3, pp. 1237–1242. doi 10.1016/j.neuroimage.2006.04.226
Arahata, K., Ishihara, T., Fukunaga, H., et al., Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses, Muscle Nerve Suppl., 1995, vol. 2, pp. S56–S66.
Frisullo, G., Frusciante, R., Nociti, V., et al., CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI, J. Clin. Immunol., 2011, vol. 31, no. 2, pp. 155–166. doi 10.1007/s10875-010-9474-6
Papa, S., Guerrieri, F., Zanotti, F., et al., F0 and F1 subunits involved in the gate and coupling function of mitochondrial H+ ATP synthase, Ann. N.Y. Acad. Sci., 1992, vol. 671, pp. 345–358.
Upadhyaya, M., Lunt, P.W., Sarfarazi, M., et al., DNA marker applicable to presymptomatic and prenatal diagnosis of facioscapulohumeral disease, Lancet, 1990, vol. 336, no. 8726, pp. 1320–1321.
Wijmenga, C., Frants, R.R., Brouwer, O.F., et al., Location of facioscapulohumeral muscular dystrophy gene on chromosome 4, Lancet, 1990, vol. 336, no. 8716, pp. 651–653.
Wijmenga, C., Padberg, G.W., Moerer, P., et al., Mapping of facioscapulohumeral muscular dystrophy gene to chromosome 4q35-qter by multipoint linkage analysis and in situ hybridization, Genomics, 1991, vol. 9, no. 4, pp. 570–575.
Mathews, K.D., Mills, K.A., Bosch, E.P., et al., Linkage localization of facioscapulohumeral muscular dystrophy (FSHD) in 4q35, Am. J. Hum. Genet., 1992, vol. 51, no. 2, pp. 428–431.
Upadhyaya, M., Lunt, P., Sarfarazi, M., et al., The mapping of chromosome 4q markers in relation to facioscapulohumeral muscular dystrophy (FSHD), Am. J. Hum. Genet., 1992, vol. 51, no. 2, pp. 404–410.
Wijmenga, C., Hewitt, J.E., Sandkuijl, L.A., et al., Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy, Nat. Genet., 1992, vol. 2, no. 1, pp. 26–30. doi 10.1038/ng0992-26
Gabriels, J., Beckers, M.C., Ding, H., et al., Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element, Gene, 1999, vol. 236, no. 1, pp. 25–32.
Deidda, G., Cacurri, S., Grisanti, P., et al., Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter, Eur. J. Hum. Genet., 1995, vol. 3, no. 3, pp. 155–167.
Butz, M., Koch, M.C., Muller-Felber, W., et al., Facioscapulohumeral muscular dystrophy: phenotypegenotype correlation in patients with borderline D4Z4 repeat numbers, J. Neurol., 2003, vol. 250, no. 8, pp. 932–937. doi 10.1007/s00415-003-1116-y
Lunt, P.W., 44th ENMC International Workshop: Facioscapulohumeral Muscular Dystrophy: Molecular Studies 19–21 July 1996, Naarden, The Netherlands, Neuromuscular Disord., 1998, vol. 8, no. 2, pp. 126–130.
Bakker, E., Wijmenga, C., Vossen, R.H., et al., The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter, Muscle Nerve Suppl., 1995, vol. 2, pp. S39–S44.
Lemmers, R.J., van der Maarel, S.M., van Deutekom, J.C., et al., Inter- and intrachromosomal sub-telomeric rearrangements on 4q35: implications for facioscapulohumeral muscular dystrophy (FSHD) aetiology and diagnosis, Hum. Mol. Genet., 1998, vol. 7, no. 8, pp. 1207–1214.
Lemmers, R.J., Wohlgemuth, M., van der Gaag, K.J., et al., Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy, Am. J. Hum. Genet., 2007, vol. 81, no. 5, pp. 884–894. doi 10.1086/521986
Lemmers, R.J., van der Vliet, P.J., van der Gaag, K.J., et al., Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution, Am. J. Hum. Genet., 2010, vol. 86, no. 3, pp. 364–377. doi 10.1016/j.ajhg.2010.01.035
Wijmenga, C., Frants, R.R., Hewitt, J.E., et al., Molecular genetics of facioscapulohumeral muscular dystrophy, Neuromuscular Disord., 1993, vol. 3, nos. 5–6, pp. 487–491.
Tupler, R., Berardinelli, A., Barbierato, L., et al., Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy, J. Med. Genet., 1996, vol. 33, no. 5, pp. 366–370.
Hewitt, J.E., Lyle, R., Clark, L.N., et al., Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy, Hum. Mol. Genet., 1994, vol. 3, no. 8, pp. 1287–1295.
Snider, L., Asawachaicharn, A., Tyler, A.E., et al., RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy, Hum. Mol. Genet., 2009, vol. 18, no. 13, pp. 2414–2430. doi 10.1093/hmg/ddp180
Dixit, M., Ansseau, E., Tassin, A., et al., DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 104, no. 46, pp. 18157–18162. doi 10.1073/pnas.0708659104
van Geel, M., Dickson, M.C., Beck, A.F., et al., Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin, Genomics, 2002, vol. 79, no. 2, pp. 210–217. doi 10.1006/geno. 2002.6690
Lejeune, E. and Allshire, R.C., Common ground: small RNA programming and chromatin modifications, Curr. Opin. Cell. Biol., 2011, vol. 23, no. 3, pp. 258–265. doi 10.1016/j.ceb.2011.03.005
Pal-Bhadra, M., Leibovitch, B.A., Gandhi, S.G., et al., Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery, Science, 2004, vol. 303, no. 5658, pp. 669–672. doi 10.1126/science.1092653
Probst, A.V., Okamoto, I., Casanova, M., et al., A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development, Dev. Cell, 2010, vol. 19, no. 4, pp. 625–638. doi 10.1016/j.devcel.2010.09.002
Sabin, L.R., Delas, M.J., and Hannon, G.J., Dogma derailed: the many influences of RNA on the genome, Mol Cell, 2013, vol. 49, no. 5, pp. 783–794. doi 10.1016/j.molcel.2013.02.010
Volpe, T.A., Kidner, C., Hall, I.M., et al., Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi, Science, 2002, vol. 297, no. 5588, pp. 1833–1837. doi 10.1126/science.1074973
Snider, L., Geng, L.N., Lemmers, R.J., et al., Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene, PLoS Genet., 2010, vol. 6, no. 10. e1001181. doi 10.1371/journal.pgen.1001181
Cabianca, D.S., Casa, V., Bodega, B., et al., A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy, Cell, 2012, vol. 149, no. 4, pp. 819–831. doi 10.1016/j.cell.2012.03.035
Gregory, G.D., Vakoc, C.R., Rozovskaia, T., et al., Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes, Mol. Cell. Biol., 2007, vol. 27, no. 24, pp. 8466–8479. doi 10.1128/MCB.00993-07
Tanaka, Y., Katagiri, Z., Kawahashi, K., et al., Trithorax- group protein ASH1 methylates histone H3 lysine 36, Gene, 2007, vol. 397, nos. 1–2, pp. 161–168. doi 10.1016/j.gene.2007.04.027
Yuan, W., Xu, M., Huang, C., et al., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation, J. Biol. Chem., 2011, vol. 286, no. 10, pp. 7983–7989. doi 10.1074/jbc.M110.194027
Tassin, A., Laoudj-Chenivesse, D., Vanderplanck, C., et al., DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy?. J. Cell. Mol. Med., 2013, vol. 17, no. 1, pp. 76–89. doi 10.1111/j.1582-4934.2012.01647.x
Bourque, G., Leong, B., Vega, V.B., et al., Evolution of the mammalian transcription factor binding repertoire via transposable elements, Genome Res., 2008, vol. 18, no. 11, pp. 1752–1762. doi 10.1101/gr.080663.108
Leidenroth, A. and Hewitt, J.E., A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene, BMC Evol. Biol., 2010, vol. 10, p. 364. doi 10.1186/1471-2148-10- 364
van Overveld, P.G., Lemmers, R.J., Sandkuijl, L.A., et al., Hypomethylation of D4Z4 in 4q-linked and non- 4q-linked facioscapulohumeral muscular dystrophy, Nat. Genet., 2003, vol. 35, no. 4, pp. 315–317. doi 10.1038/ng1262
de Greef, J.C., Lemmers, R.J., van Engelen, B.G., et al., Common epigenetic changes of D4Z4 in contraction- dependent and contraction-independent FSHD, Hum. Mutat., 2009, vol. 30, no. 10, pp. 1449–1459. doi 10.1002/humu.21091
Zeng, W., de Greef, J.C., Chen, Y.Y., et al., Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD), PLoS Genet., 2009, vol. 5, no. 7. e1000559. doi 10.1371/journal.pgen.1000559
Balog, J., Thijssen, P.E., de Greef, J.C., et al., Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD, Epigenetics, 2012, vol. 7, no. 6, pp. 579–584. doi 10.4161/epi.20001
Kowaljow, V., Marcowycz, A., Ansseau, E., et al., The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein, Neuromuscular Disord., 2007, vol. 17, no. 8, pp. 611–623. doi 10.1016/j.nmd.2007.04.002
Wallace, L.M., Garwick, S.E., Mei, W., et al., DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo, Ann. Neurol., 2011, vol. 69, no. 3, pp. 540–552. doi 10.1002/ana.22275
Lemmers, R.J., Tawil, R., Petek, L.M., et al., Digenic inheritance of an SMCHD1 mutation and an FSHDpermissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2, Nat. Genet., 2012, vol. 44, no. 12, pp. 1370–1374. doi 10.1038/ng.2454
Blewitt, M.E., Gendrel, A.V., Pang, Z., et al., SmcHD1, containing a structural-maintenance-ofchromosomes hinge domain, has a critical role in X inactivation, Nat. Genet., 2008, vol. 40, no. 5, pp. 663–669. doi 10.1038/ng.142
Gendrel, A.V., Apedaile, A., Coker, H., et al., Smchd1- dependent and-independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome, Dev. Cell, 2012, vol. 23, no. 2, pp. 265–279. doi 10.1016/j.devcel.2012.06.011
Gendrel, A.V., Tang, Y.A., Suzuki, M., et al., Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes, Mol. Cell. Biol., 2013, vol. 33, no. 16, pp. 3150–3165. doi 10.1128/MCB.00145-13
Mould, A.W., Pang, Z., Pakusch, M., et al., Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation, Epigenet. Chromatin, 2013, vol. 6, no. 1, pp. 19. doi 10.1186/1756-8935-6-19
Larsen, M., Rost, S., El Hajj, N., et al., Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1, Eur. J. Hum. Genet., 2015, vol. 23, no. 6, pp. 808–816. doi 10.1038/ejhg.2014.191
Lunt, P.W., Compston, D.A., and Harper, P.S., Estimation of age dependent penetrance in facioscapulohumeral muscular dystrophy by minimising ascertainment bias, J. Med. Genet., 1989, vol. 26, no. 12, pp. 755–760.
Scionti, I., Greco, F., Ricci, G., et al., Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy, Am. J. Hum. Genet., 2012, vol. 90, no. 4, pp. 628–635. doi 10.1016/j.ajhg.2012.02.019
Statland, J.M., McDermott, M.P., Heatwole, C., et al., Reevaluating measures of disease progression in facioscapulohumeral muscular dystrophy, Neuromuscular Disord., 2013, vol. 23, no. 4, pp. 306–312. doi 10.1016/j.nmd.2013.01.008
The FSH-DY Group, A prospective, quantitative study of the natural history of facioscapulohumeral muscular dystrophy (FSHD): implications for therapeutic trials, Neurology, 1997, vol. 48, no. 1, pp. 38–46.
Busse, K., Kohler, J., Stegmann, K., et al., An inherited 4q35-EcoRI-DNA-fragment of 35 kb in a family with a sporadic case of facioscapulohumeral muscular dystrophy (FSHD), Neuromuscular Disord., 2000, vol. 10, no. 3, pp. 178–181.
Tsumagari, K., Chen, D., Hackman, J.R., et al., FSH dystrophy and a subtelomeric 4q haplotype: a new assay and associations with disease, J. Med. Genet., 2010, vol. 47, no. 11, pp. 745–751. doi 10.1136/jmg.2009.076703
de Greef, J.C., Frants, R.R., and van der Maarel, S.M., Epigenetic mechanisms of facioscapulohumeral muscular dystrophy, Mutat. Res., 2008, vol. 647, nos. 1–2, pp. 94–102. doi 10.1016/j.mrfmmm.2008.07.011
Jones, T.I., Yan, C., Sapp, P.C., et al., Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing, Clin. Epigenet., 2014, vol. 6, no. 1, p. 23. doi 10.1186/1868-7083-6-23
Nguyen, K., Walrafen, P., Bernard, R., et al., Molecular combing reveals allelic combinations in facioscapulohumeral dystrophy, Ann. Neurol., 2011, vol. 70, no. 4, pp. 627–633. doi 10.1002/ana.22513
Vasale, J., Boyar, F., Jocson, M., et al., Molecular combing compared to Southern blot for measuring D4Z4 contractions in FSHD, Neuromuscular Disord., 2015, vol. 25, no. 12, pp. 945–951. doi 10.1016/j.nmd. 2015.08.008
Newlands, S., Levitt, L.K., Robinson, C.S., et al., Transcription occurs in pulses in muscle fibers, Genes Dev., 1998, vol. 12, no. 17, pp. 2748–2758.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © N.V. Zernov, A.V. Marakhonov, J.V. Vyakhireva, A.A. Guskova, E.L. Dadali, M.Yu. Skoblov, 2017, published in Genetika, 2017, Vol. 53, No. 6, pp. 651–662.
Rights and permissions
About this article
Cite this article
Zernov, N.V., Marakhonov, A.V., Vyakhireva, J.V. et al. Clinical and genetic characteristics and diagnostic features of Landouzy–Dejerine facioscapulohumeral muscular dystrophy. Russ J Genet 53, 640–650 (2017). https://doi.org/10.1134/S102279541706014X
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102279541706014X