Abstract
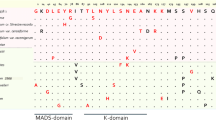
FRUITFULL (FUL) transcription factors, SlFUL1 and SlFUL2, are directly involved in the regulation of tomato fruit development. In the present study, complete sequences of novel SlFUL2 orthologous genes were isolated and structurally characterized in five accessions of domestic tomato Solanum lycopersicum and closely related wild species S. pimpinellifolium, having different morphological characteristics. In the studied genes, overall level of nucleotide sequence variation was 1.94%. Nine out of 11 exon-specific SNPs led to amino acid substitutions in the functionally important MADS, K, and C domains of the SlFUL2 orthologues. Considerable differences in the level and spatiotemporal dynamics of the expression patterns in different tomato organs at the species and intraspecific levels were observed.
Similar content being viewed by others
References
Peralta, I.E. and Spooner, D.M., History, origin and early cultivation of tomato (Solanaceae), in Genetic Improvement of Solanaceous Crops, vol. 2: Tomato, Enfield: Science Publishers, 2006, pp. 1–27.
Chetelat, R.T. and Ji, Y., Cytogenetics and evolution, in Genetic Improvement of Solanaceous Crops, vol. 2: Tomato, Enfield: Science Publishers, 2006, pp. 77–112.
Theißen, G., Development of floral organ identity: stories from the MADS house, Curr. Opin. Plant Biol., 2001, vol. 4, pp. 75–85.
Bemer, M., Karlova, R., Ballester, A.R., et al., The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening, Plant Cell, 2012, vol. 24, no. 11, pp. 4437–4451. doi 10.1105/tpc.112.103283
Ito, Y., Regulation of tomato fruit ripening by MADSbox transcription factors, JARQ, 2016, vol. 50, no. 1, pp. 33–38.
Fujisawa, M., Shima, Y., Nakagawa, H., et al., Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins, Plant Cell, 2014, vol. 26, no. 1, pp. 89–101. doi 10.1105/tpc.113.119453
Wang, S., Lu, G., Hou, Z., et al., Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening, J. Exp. Bot., 2014, vol. 65, no. 12, pp. 3005–3014. doi 10.1093/jxb/eru137
Kochieva, E.Z. and Suprunova, T.P., Identification of inter- and intraspecific polymorphism in tomato, Russ. J. Genet., 1999, vol. 35, no. 10, pp. 1194–1196.
Tamura, K., Stecher, G., Peterson, D., et al., MEGA6: molecular evolutionary genetics analysis version 6.0, Mol. Biol. Evol., 2013, vol. 30, pp. 2725–2729. doi 10.1093/molbev/mst197
GraphPad Prism version 7.02 for Windows, San Diego, California: GraphPad Software. http://www.graphpad. com.
Vandenbussche, M., Theissen, G., Van de Peer, Y., et al., Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations, Nucleic Acids Res., 2003, vol. 31, pp. 4401–4409.
Parenicová, L., de Folter, S., Kieffer, M., et al., Molecular and phylogenetic analyses of the complete MADSbox transcription factor family in Arabidopsis: new openings to the MADS world, Plant Cell, 2003, vol. 15, pp. 1538–1551.
Lutova, L.A., Ezhova, T.A., Dodueva, I.E., and Osipova, M.A., Genetic control of plant development, in Genetika razvitiya rastenii (Genetics of Plant Development), St. Petersburg: N-L, 2010.
Santelli, E. and Richmond, T.J., Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution, J. Mol. Biol., 2000, vol. 297, no. 2, pp. 437–449.
Kaufmann, K., Melzer, R., and Theissen, G., MIKCtype MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants, Gene, 2005, vol. 347, pp. 183–198.
Hittinger, C. and Carroll, S., Gene duplication and the adaptive evolution of a classic genetic switch, Nature, 2007, vol. 449, pp. 677–682.
Litt, A. and Irish, V., Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development, Genetics, 2003, vol. 165, pp. 821–833.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © M.A. Slugina, A.V. Shchennikova, E.Z. Kochieva, 2017, published in Genetika, 2017, Vol. 53, No. 6, pp. 687–695.
Rights and permissions
About this article
Cite this article
Slugina, M.A., Shchennikova, A.V. & Kochieva, E.Z. Novel SlFUL2 orthologous genes and analysis of their expression in wild and cultivated tomato of the section Lycopersicon. Russ J Genet 53, 672–679 (2017). https://doi.org/10.1134/S1022795417060126
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795417060126