Abstract
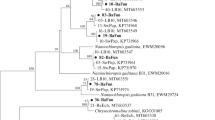
The diversity of the symbiotic community of the endemic Baikal sponge Swartschewskia papyracea was studied, and an analysis of the polyketide synthases genes spectrum in sponge-associated microorganisms was carried out. Six bacterial phyla were detected in the S. papyracea microbiome: Verrucomicrobia, Cyanobacteria, Actinobacteria, Bacteroidetes, Proteobacteria, and Planctomycetes. Unlike the microbial associations of other freshwater sponges, the community under study was dominated by the phylaVerrucomicrobia (42.1%) and Cyanobacteria (17.5%), while the proportion of the Proteobacteria was unusually low (9.7%). In the S. papyracea community metagenome, there were identified 18 polyketide synthases genes fragments, the closest homologues of which included the polyketide synthases of the microorganisms belonging to the bacterial phyla Cyanobacteria, Proteobacteria (classes Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria), and Acidobacteria as well as the eukaryotic algae of the phylum Heterokonta (class Eustigmatophyceae). Polyketide synthase sequences from S. papyracea formed three groups on the phylogenetic tree: a group of hybrid NRPS/PKS complexes, a group of cyanobacterial polyketide synthases, and a group of homologues of the eukaryotic alga Nannochloropsis gaditana. Notably, the identified polyketide synthase genes fragments showed only a 57–88% similarity to the sequences from the databases, which implies the presence of genes controlling the synthesis of the novel, still unstudied, polyketide compounds in the S. papyracea community. It was proposed that the habitat conditions of S. papyracea affect the taxonomic composition of the microorganisms associated with the sponge, including the diversity of the producers of secondary metabolites.
Similar content being viewed by others
References
Webster, N.S. and Taylor, M.W., Marine sponges and their microbial symbionts: love and other relationships, Environ. Microbiol., 2012, vol. 14, no. 2, pp. 335–346.
Thomas, T.R., Kavlekar, D.P., and LokaBharathi, P.A., Marine drugs from sponge microbe association—a review, Mar. Drugs, 2010, vol. 8, pp. 1417–1468.
Efremova, S.M., Sponges, in Annotirovannyi spisok fauny ozera Baikal i ego vodosbornogo basseina (An Annotated List of the Fauna of Lake Baikal and Its Catchment Area), Timoshkin, O.A., Ed., Novosibirsk: Nauka, 2001, vol. 1, pp. 177–190.
Kozhov, M.M., Biologiya ozera Baikal (Biology of Lake Baikal), Moscow: Akad. Nauk SSSR, 1962.
Kaluzhnaya, O.V., Krivich, A.A., and Itskovich, V.B., Diversity of 16S rRNA genes in metagenomic community of the freshwater sponge Lubomirskia baicalensis, Russ. J. Genet., 2012, vol. 48, no. 8, pp. 855–858.
Kaluzhnaya, O.V. and Itskovich, V.B., Phylogenetic diversity of microorganisms associated with the deepwater sponge Baikalospongia intermedia, Russ. J. Genet., 2014, vol. 50, no. 7, pp. 667–676.
Gladkikh, A.S., Kaluzhnaya, O.V., Belykh, O.I., et al., Analysis of bacterial communities of two Lake Baikal endemic sponge species, Microbiology (Moscow), 2014, vol. 83, no. 6, pp. 787–797.
Kaluzhnaya, O.V., Kulakova, N.V., and Itskovich, V.B., Diversity of polyketide synthase (PKS) genes in metagenomic community of freshwater sponge Lubomirskia baicalensis, Mol. Biol. (Moscow), 2012, vol. 46, no. 6, pp. 790–795.
Kaluzhnaya, O.V., Lipko, I.A., Itskovich, V.B., et al., PCR-screening of bacteria isolated from freshwater sponge Lubomirskia baicalensis for detection of genes of secondary metabolites, Voda: Khim. Ekol., 2013, no. 7, pp. 70–74.
Lipko (Terkina), I.A., Kaluzhnaya, O.V., Kravchenko, O.S., and Parfenova, V.V., Identification of polyketide synthase genes in genome of Pseudomonas fluorescens strain 28Bb-06 from freshwater sponge Baikalospongia bacillifera, Mol. Biol. (Moscow), 2012, vol. 46, no. 4. pp. 609–611.
Schröder, H.C., Efremova, S.M., Itskovich, V.B., et al., Molecular phylogeny of the freshwater sponges in Lake Baikal, J. Zool. Syst. Evol. Res., 2003, vol. 41, pp. 80–86.
Itskovich, V., Gontcharov, A., Masuda, Y., et al., Ribosomal ITS sequences allow resolution of freshwater sponge phylogeny with alignments guided by secondary structure prediction, J. Mol. Evol., 2008, vol. 67, pp. 608–620.
Rezvoi, P.D., Prenovodnye gubki (Freshwater Sponges), vol. 2, no. 2 of Fauna SSSR: gubki (Fauna of the Soviet Union: Sponges), Zernov S.A., Ed., Moscow: Akad. Nauk SSSR, 1936.
Masuda, Y., Studies on the taxonomy and distribution of freshwater sponges in Lake Baikal, Prog. Mol. Subcell. Biol., 2009, vol. 47, pp. 81–110.
Timoshkin, O.A., Atlas i opredelitel’ pelagobiontov Baikala (s kratkimi ocherkami po ikh ekologii) (Atlas and Identification Book of Pellagobionts of Lake Baikal (with Brief Essays on Their Ecology)), Timoshkin, O.A., Mazepova, G.F., Mel’nik, N.G., et al., Eds., Novosibirsk: Nauka, 1995.
Pile, A.J., Patterson, M.R., Savarese, M., et al., Trophic effects of sponge feeding within Lake Baikal’s littoral zone: 2. Sponge abundance, diet, feeding efficiency, and carbon flux, Limnol. Oceonogr., 1997, vol. 42, no. 1, pp. 178–184.
Hochmuth, T. and Piel, J., Polyketide synthases of bacterial symbionts in sponges—evolution-based applications in natural products research, Phytochemistry, 2009, vol. 70, nos. 15–16, pp. 1841–1849.
Esteves, A.I., Hardoim, C.C., Xavier, J.R., et al., Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the northeast Atlantic, FEMS Microbiol. Ecol., 2013, vol. 85, no. 3, pp. 519–536.
Della Sala, G., Hochmuth, T., Teta, R., et al., Polyketide synthases in the microbiome of the marine sponge Plakortis halichondrioides: a metagenomic update, Mar. Drugs, 2014, vol. 12, pp. 5425–5440.
Santos, O.C., Soares, A.R., Machado, F.L., et al., Investigation of biotechnological potential of spongeassociated bacteria collected in Brazilian coast, Lett. Appl. Microbiol., 2015, vol. 60, no. 2, pp. 140–147.
Jenke-Kodama, H. and Dittmann, E., Evolution of metabolic diversity: insights from microbial polyketide synthases, Phytochem., 2009, vol. 70, pp. 1858–1866.
Barrios-Llerena, M.E., Burja, A.M., and Wright, P.C., Genetic analysis of polyketide synthase and peptide synthetase genes in cyanobacteria as a mining tool for secondary metabolites, J. Ind. Microbiol. Biotechnol., 2007, vol. 34, pp. 443–456.
Hall, T.A., BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT, Nucleic Acids. Symp., 1999, vol. 41, pp. 95–98.
Ashelford, K.E., Chuzhanova, N.A., Fry, J.C., et al., New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras, Appl. Environ. Microbiol., 2006, vol. 72, no. 9, pp. 5734–5741.
Altschul, S.F., Warren, G., Miller, W., et al., Basic local alignment search tool, J. Mol. Biol., 1990, vol. 215, pp. 403–410.
Tamura, K., Dudley, J., Nei, M., and Kumar, S., MEGA4: molecular genetics analysis (MEGA) software version 4.0, Mol. Biol. Evol., 2007, vol. 24, pp. 1596–1599.
Newton, R.J., Jones, S.E., Eiler, A., et al., A guide to the natural history of freshwater lake bacteria, Microb. Mol. Biol. Rev., 2011, vol. 75, no. 1, pp. 14–49.
Lee, K.-C., Webb, R.I., Janssen, P.H., et al., Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes, BMC Microbiol., 2009, vol. 9, no. 5. http://www.biomedcentral.com/1471-2180/9/5
Cardman, Z., Arnosti, C., Durbin, A., et al., Verrucomicrobia are candidates for polysaccharide-degrading bacterioplankton in an Arctic fjord of Svalbard, Appl. Environ. Microbiol., 2014, vol. 80, no. 12, pp. 3749–3756.
Zhang, J., Zhang, X., Liu, Y., et al., Bacterioplankton communities in a high-altitude freshwater wetland, Ann. Microbiol., 2014, vol. 64, no. 3, pp. 1405–1411.
Parfenova, V.V., Gladkikh, A.S., and Belykh, O.I., Comparative analysis of biodiversity in the planktonic and biofilm bacterial communities in Lake Baikal, Microbiology (Moscow), 2013, vol. 82, no. 1, pp. 91–101.
Costa, R., Keller-Costa, T., Gomes, N.C.M., et al., Evidence for selective bacterial community structuring in the freshwater sponge Ephydatia fluviatilis, Microb. Ecol., 2013, vol. 65, pp. 232–244.
King, G.M., Smith, C.B., Tolar, B., and Hollibaugh, J.T., Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the northern Gulf of Mexico, Front. Microbiol., 2013, vol. 3, no. 438. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3548560/
Arnds, J., Knittel, K., Buck, U., et al., Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake, Syst. Appl. Microbiol., 2010, vol. 33, pp. 139–148.
Gernert, C., Glockner, F.O., Krohne, G., et al., Microbial diversity of the freshwater sponge Spongilla lacustris, Microb. Ecol., 2005, vol. 50, pp. 206–212.
Belykh, O.I., Pomazkina, G.V., Tikhonova, I.V., and Tomberg, I.V., Characteristic of summer phytoplankton and autotrophic picoplankton of Lake Baikal in 2005, Algologiya, 2007, vol. 17, pp. 380–396.
Lemloh, M.-L., Fromont, J., Brümmer, F., and Usher, K.L., Diversity and abundance of photosynthetic sponges in temperate Western Australia, BMC Ecol., 2009, vol. 9, no. 4. http://www.biomedcentral.com/1472-6785/9/4
Erwin, P.M., López-Legentil, S., Gonzalez-Pech, R., and Turon, X., A specific mix of generalists: bacterial symbionts in mediterranean Ircinia spp., FEMS Microb. Ecol., 2012, vol. 79, no. 3, pp. 619–637.
Ghai, R., Mizuno, C.M., Picazo, A., et al., Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing, Mol. Ecol., 2014, vol. 23, no. 24, pp. 6073–6090.
Jezbera, J., Sharma, A.K., Brandt, U., et al., “Candidatus Planktophila limnetica,” an actinobacterium representing one of the most numerically important taxa in freshwater bacterioplankton, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, pp. 2864–2869.
Karlov, D.S., Mari, D., Chuvochina, M.S., et al., Microbial communities of water column of Lake Radok, East Antarctica, dominated by abundant actinobacterium “Candidatus Planktophila limnetica”, Microbiology (Moscow), 2011, vol. 80, no. 4, pp. 576–579.
Zwart, G., Crump, B., Agterveld, M., et al., Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers, Aquat. Microb. Ecol., 2002, vol. 28, pp. 141–155.
Qu, J.-H. and Yuan, H.-L., Sediminibacterium salmoneum gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from sediment of a eutrophic reservoir, Int. J. Syst. Evol. Microbiol., 2008, vol. 58, pp. 2191–2194.
Fernandez-Gomez, B., Richter, M., Schüler, M., et al., Ecology of marine Bacteroidetes: a comparative genomics approach, ISME J., 2013, vol. 7, pp. 1026–1037.
Wu, J., Gao, W., Johnson, R.H., et al., Integrated metagenomic and metatranscriptomic analyses of microbial communities in the mesoand bathypelagic realm of North Pacific Ocean, Mar. Drugs, 2013, vol. 11, pp. 3777–3801.
Villaescusa, J.A., Casamayora, E.O., Rochera, C., et al., Heterogeneous vertical structure of the bacterioplankton community in a non-stratified Antarctic lake, Antarct. Sci., 2013, vol. 25, no. 2, pp. 229–238.
Taylor, M.W., Radax, R., Steger, D., and Wagner, M., Sponge-associated microorganisms: evolution, ecology, and biotechnological potential, Microbiol. Mol. Biol. Rev., 2007, vol. 71, no. 2, pp. 295–347.
Kasalicky V., Jezbera J., Hahn M.W., and Šimek K., The diversity of the Limnohabitans genus, an important group of freshwater bacterioplankton, by characterization of 35 isolated strains, PLoS One, 2013, vol. 8, no. 3. e58209. doi 10.1371/journal.pone.0058209
Jezbera, J., Jezberova, J., Kasalicky, V., et al., Patterns of Limnohabitans microdiversity across a large set of freshwater habitats as revealed by reverse line blot hybridization, PLoS One, 2013, vol. 8, no. 3. e58527. doi 10.1371/journal.pone.0058527
Paver, S.F., Hayek, K.R., Gano, K.A., et al., Interactions between specific phytoplankton and bacteria affect lake bacterial community succession, Environ. Microbiol., 2013, vol. 15, no. 9, pp. 2489–2504.
Ehrenreich, I., Waterbury, J., and Webb, E., Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes, Appl. Envir. Microb., 2005, vol. 71, pp. 7401–7413.
Edwards, D.J., Marquez, B.L., Nogle, L.M., et al., Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscule, Chem. Biol., 2004, vol. 11, no. 6, pp. 817–833.
Forli, S., Epothilones: from discovery to clinical trials, Curr. Top. Med. Chem., 2014, vol. 14, no. 20, pp. 2312–2321.
Khosla, C., Tang, Y., Chen, A.Y., et al., Structure and mechanism of the 6-deoxyerythronolide B synthase, Annu. Rev. Biochem., 2007, vol. 76, pp. 195–221.
Wang, D., Ning, K., Li, J., et al., Nannochloropsis genomes reveal evolution of microalgal oleaginous traits, PLoS Genet., 2014, vol. 10, no. 1. e1004094. doi 10.1371/journal.pgen.1004094
Szaby M., Parker, K., Guruprasad, S., et al., Photosynthetic acclimation of Nannochloropsis oculata investigated by multi-wavelength chlorophyll fluorescence analysis, Bioresour. Technol., 2014, vol. 167, pp. 521–529.
Fietz, S., Bleiß, W., Hepperle, D., et al., First record of Nannochloropsis limnetica (Eustigmatophyceae) in the autotrophic phytoplankton from Lake Baikal, J. Phycol., 2005, vol. 41, no. 4, pp. 780–790.
Tamburic, B., Szaby M., Tran, N.-A.T., et al., Action spectra of oxygen production and chlorophyll a fluorescence in the green microalga Nannochloropsis oculata, Bioresour. Technol., 2014, vol. 169, pp. 320–327.
Konga, W., Reamb, D.C., Priscuc, J.C., and MorganKissb, R.M., Diversity and expression of RubisCO genes in a perennially ice-covered Antarctic lake during the polar night transition, Appl. Environ. Microbiol., 2012, vol. 78, no. 12, pp. 4358–4366.
Simionato, D., Block, M.A., La Rocca, N., et al., The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus, Eukaryot. Cell, 2013, vol. 12, no. 5, pp. 665–676.
Starkenburg, S., Kwon, K.J., Jha, R.K., et al., A pangenomic analysis of the Nannochloropsis organellar genomes reveals novel genetic variations in key metabolic genes, BMC Genomics, 2014, vol. 15, no. 212. http://www.biomedcentral.com/1471-2164/15/212
Krienitz, L., Hepperle, D., Stich, H.-B., and Weiler, W., Nannochloropsis limnetica (Eustigmatophyceae), a new species of picoplankton from freshwater, Phycologia, 2000, vol. 39, pp. 219–227.
Fawley, K.P. and Fawley, M.W., Observations on the diversity and ecology of freshwater Nannochloropsis (Eustigmatophyceae), with descriptions of new taxa, Protist, 2007, vol. 158, no. 3, pp. 325–336.
Kehr, J.-C., Picchi, D.G., Dittmann, E., et al., Natural product biosyntheses in cyanobacteria: a treasure trove of unique enzymes, Beilstein. J. Org. Chem., 2011, vol. 7, pp. 1622–1635.
Bil, K., Titlyanov, E., Berner, T., et al., Some aspects of the physiology and biochemistry of Lubomirskia baikalensis, a sponge from Lake Baikal containing symbiotic algae, Symbiosis, 1999, vol. 26, pp. 179–191.
Sukenik, A., Beardall, J., Kromkamp, J.C., et al., Photosynthetic performance of outdoor Nannochloropsis mass cultures under a wide range of environmental conditions, Aquat. Microb. Ecol., 2009, vol. 56, pp. 297–308.
Vinyard, D.J., Gimpel, J., Ananyev, G.M., et al., Natural variants of photosystem II subunit D1 tune photochemical fitness to solar intensity, J. Biol. Chem., 2013, vol. 288, no. 8, pp. 5451–5462.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © O.V. Kaluzhnaya, V.B. Itskovich, 2016, published in Genetika, 2016, Vol. 52, No. 1, pp. 47–58.
Rights and permissions
About this article
Cite this article
Kaluzhnaya, O.V., Itskovich, V.B. Distinctive features of the microbial diversity and the polyketide synthase genes spectrum in the community of the endemic Baikal sponge Swartschewskia papyracea . Russ J Genet 52, 38–48 (2016). https://doi.org/10.1134/S1022795416010099
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795416010099