Abstract
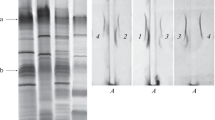
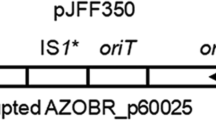
In the bacterium Azospirillum brasilense Sp245, extracellular calcofluor-binding polysaccharides (Cal+ phenotype) and two types of lipopolysaccharides, LPSI and LPSII, were previously identified. These lipopolysaccharides share the same repeating O-polysaccharide unit but have different antigenic structures and different charges of their O-polysaccharides and/or core oligosaccharides. Several dozens of predicted genes involved in the biosynthesis of polysaccharides have been localized in the AZOBR_p6 plasmid of strain Sp245 (GenBank accession no. HE577333). In the present work, it was demonstrated that an artificial transposon Omegon-Km had inserted into the central region of the AZOBR_p60120 gene in the A. brasilense Sp245 LPSI− Cal− KM252 mutant. In A. brasilense strain Sp245, this plasmid gene encodes a putative glycosyltransferase containing conserved domains characteristic of the enzymes participating in the synthesis of O-polysaccharides and capsular polysaccharides (accession no. YP004987664). In mutant KM252, a respective predicted protein is expected to be completely inactivated. As a result of the analysis of the EcoRI fragment of the AZOBR_p6 plasmid, encompassing the AZOBR_p60120 gene and a number of other loci, novel data on the structure of AZOBR_p6 were obtained: an approximately 5-kb gap (GenBank accession no. KM189439) was closed in the nucleotide sequence of this plasmid.
Similar content being viewed by others
References
Katsy, E.I., Molekulyarnaya genetika assotsiativnogo vzaimodeistviya bakterii i rastenii: sostoyanie i perspektivy issledovanii, (Molecular Genetics of Associative Interaction between Bacteria and Plants: Current Status and Prospects of the Studies), Ignatov, V.V., Ed., Moscow: Nauka, 2007.
Baldani, V.L.D., Baldani, J.I., and Döbereiner, J., Effects of Azospirillum inoculation on root infection and nitrogen incorporation in wheat, Can. J. Microbiol., 1983, vol. 29, no. 8, pp. 924–929.
Katsy, E.I., Plasmid rearrangements and changes in cell-surface architecture and social behavior of Azospirillum brasilense, in Plasticity in Plant-Growth-Promoting and Phytopathogenic Bacteria, Katsy, E.I., Ed., New York: Springer, 2014, pp. 81–97.
Katzy, E.I., Matora, L.Yu., Serebrennikova, O.B., and Scheludko, A.V., Involvement of a 120-MDa plasmid of Azospirillum brasilense Sp245 in production of lipopolysaccharides, Plasmid, 1998, vol. 40, no. 1, pp. 73–83.
Fedonenko, Yu.P., Zdorovenko, E.L., Konnova, S.A., et al., A comparison of the lipopolysaccharides and O-specific polysaccharides of Azospirillum brasilense Sp245 and its Omegon-Km mutants KM018 and KM252, Microbiology (Moscow), 2004, vol. 73, no. 2, pp. 143–149.
Fedonenko, Y.P., Zatonsky, G.V., Konnova, S.A., et al., Structure of the O-specific polysaccharide of the lipopolysaccharide of Azospirillum brasilense Sp245, Carbohydr. Res., 2002, vol. 337, no. 9, pp. 869–872.
Del Gallo, M., Negi, M., and Neyra, C.A., Calcofluor and lectin-binding exocellular polysaccharides of Azospirillum brasilense and Azospirillum lipoferum, J. Bacteriol., 1989, vol. 171, no. 6, pp. 3504–3510.
Konnova, S.A., Skvortsov, I.M., Makarov, O.E., and Ignatov, V.V., Properties of polysaccharide complexes produced by Azospirillum brasilense and polysaccharides isolated from these complexes, Microbiologiya, 1994, vol. 63, no. 6, pp. 1020–1030.
Wisniewski-Dyé, F., Borziak, K., Khalsa-Moyers, G., et al., Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments, PLoS Genet., 2011, vol. 7, no. 12. e1002430.
Katsy, E.I., Borisov, I.V., Petrova, L.P., and Matora, L.Yu., The use of fragments of the 85- and 120-MDa plasmids of Azospirillum brasilense Sp245 to study the plasmid rearrangement in this bacterium and to search for homologous sequences in plasmids of Azospirillum brasilense Sp7, Russ. J. Genet., 2002, vol. 38, no. 2, pp. 124–131.
Katsy, E.I., Petrova, L.P., Kulibyakina, O.V., and Prilipov, A.G., Analysis of Azospirillum brasilense plasmid loci coding for (lipo)polysaccharides synthesis enzymes, Microbiology (Moscow), 2010, vol. 79, no. 2, pp. 216–222.
Fellay, R., Krisch, H.M., Prentki, P., and Frey, J., Omegon-Km: a transposable element designed for in vivo insertional mutagenesis and cloning of genes in gram-negative bacteria, Gene, 1989, vol. 76, no. 2, pp. 215–226.
Sambrook, J., Fritsch, E.F., and Maniatis, T., Molecular Cloning: A Laboratory Manual, New York: Cold Spring Harbor Lab., 1989, 2nd ed.
Döbereiner, J. and Day, J.M., Associative symbiosis in tropical grass: characterization of microorganisms and dinitrogen fixing sites, in Symposium on Nitrogen Fixation, Newton, W.E. and Nijmans, C.J., Eds., Pullman: Washington State Univ., 1976, pp. 518–538.
Katsy, E.I. and Prilipov, A.G., Mobile elements of an Azospirillum brasilense Sp245 85-MDa plasmid involved in replicon fusions, Plasmid, 2009, vol. 62, no. 1, pp. 22–29.
Wheeler, D.L., Church, D.M., Federhen, S., et al., Database resources of the National Center for Biotechnology, Nucleic Acids Res., 2003, vol. 31, no. 1, pp. 28–32.
Boratyn, G.M., Camacho, C., Cooper, P.S., et al., BLAST: a more efficient report with usability improvements, Nucleic Acids Res., 2013, vol. 41, pp. W29–W33.
Letunic, I., Doerks, T., and Bork, P., SMART 7: recent updates to the protein domain annotation resource, Nucleic Acids Res., 2012, vol. 40, pp. D302–D305.
Yu, N.Y., Wagner, J.R., Laird, M.R., et al., PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes, Bioinformatics, 2010, vol. 26, no. 13, pp. 1608–1615.
Juncker, A.S., Willenbrock, H., Von Heijne, G., et al., Prediction of lipoprotein signal peptides in Gram-negative bacteria, Protein Sci., 2003, vol. 12, no. 8, pp. 1652–1662.
Dyrløv Bendtsen, J., Kiemer, L., Fausböll, A., and Brunak, S., Non-classical protein secretion in bacteria, BMC Microbiol., 2005, vol. 5, p. 58.
Dyrløv Bendtsen, J., Nielsen, H., Widdick, D., et al., Prediction of twin-arginine signal peptides, BMC Bioinform., 2005, vol. 6, p. 167.
Petersen, T.N., Brunak, S., von Heijne, G., and Nielsen, H., SignalP 4.0: discriminating signal peptides from transmembrane regions, Nat. Methods, 2011, vol. 8, no. 10, pp. 785–786.
Fedonenko, Yu.P., Katsy, E.I., Petrova, L.P., et al., The structure of the O-specific polysaccharide from a mutant of nitrogen-fixing rhizobacterium Azospirillum brasilense Sp245 with an altered plasmid content, Russ. J. Bioorg. Chem., 2010, vol. 36, no. 2, pp. 219–223.
Katsy, E.I., Plasmid plasticity in the plant-associated bacteria of the genus Azospirillum, in Bacteria in Agrobiology: Plant Growth Responses, Maheshwari, D.K., Ed., Berlin: Springer, 2011, pp. 139–157.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © E.I. Katsy, A.G. Prilipov, 2015, published in Genetika, 2015, Vol. 51, No. 3, pp. 306–311.
Rights and permissions
About this article
Cite this article
Katsy, E.I., Prilipov, A.G. Insertional mutation in the AZOBR_p60120 gene is accompanied by defects in the synthesis of lipopolysaccharide and calcofluor-binding polysaccharides in the bacterium Azospirillum brasilense Sp245. Russ J Genet 51, 245–250 (2015). https://doi.org/10.1134/S1022795415030059
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795415030059