Abstract
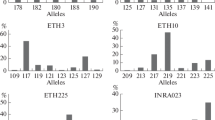
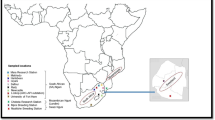
Genetic structure and diversity of 3789 animals of the Brahman breed from 23 Colombian regions were assessed. Considering the Brahman Zebu cattle as a single population, the multilocus test based on the HW equilibrium, shows significant differences (P < 0.001). Genetic characterization made on the cattle population allowed to examine the genetic variability, calculating a H o = 0.6621. Brahman population in Colombia was a small subdivision wthin populations (F it = 0.045), a geographic subdivision almost non-existent or low differentiation (F st = 0.003) and the F is calculated (0.042) indicates no detriment to the variability in the population, despite the narrow mating takes place or there is a force that causes the variability is sustained without inbreeding actually affect the cattle population. The outcomes of multivariate analyses, Bayesian inferences and interindividual genetic distances suggested that there is no genetic sub-structure in the population, because of the high rate of animal migration among regions.
Similar content being viewed by others
References
Edwards, C.J., Baird, J.F., and MacHugh, D.E., Taurine and zebu admixture in Near Eastern cattle: a comparison of mitochondrial, autosomal, and Y-chromosomal data, Anim. Genet., 2007, vol. 38, pp. 520–524.
Hansen, P.J., Physiological and cellular adaptations of zebu cattle to thermal stress, Anim. Reprod. Sci., 2004, vol. 82–83, pp. 349–360.
Meirelles, F.V., Rosa, A.J.M., Lôbo, R.B., et al., Is the zebu really Bos indicus? Genet. Mol. Biol., 1999, vol. 22, pp. 543–546.
Novoa, M.A. and Usaquén, W., Population genetic analysis of the Brahman cattle (Bos indicus) in Colombia with microsatellite markers, J. Anim. Breed. Genet., 2010, vol. 127, pp. 161–168.
Blott, S.C., Williams, J.L., Haley, C.S., Genetic relationships among European cattle breeds, Anim. Genet., 1999, vol. 29, pp. 273–282.
Cañon, J., Alexandrino, P., Bessa, I., et al., Phylogenetic analysis: models and estimation procedure, Am. J. Hum. Genet., 1967, vol. 19, pp. 233–257.
Kantanen, J., Olsaker, I., Holm, L.E., et al., Genetic diversity and population structure of 20 North European cattle breeds, J. Hered., 2000, vol. 91, pp 446–457.
Li, M.H., Tapio, I., Vilkki, J., et al., The genetic structure of cattle populations (Bos taurus) in northern Eurasia and the neighboring Near Eastern regions: implications for breeding strategies and conservation, Mol. Ecol., 2007, vol. 16, pp. 3839–3853.
MacHugh, D.E., Shriver, M.D., Loftus, R.T., et al., Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus), Genetics, 1997, vol. 146, pp. 1071–1086.
Troy, C.S., MacHugh, D.E., Bailey, J.F., et al., Genetic evidence for Near-Eastern origins of European cattle, Nature, 2001, vol. 410, pp. 1088–1091.
Freeman, A.R., Meghen, C.M., MacHugh, D.E., et al., Admixture and diversity in West African cattle populations., Mol. Ecol., 2004, vol. 13, pp. 3477–3487.
Ibeagha-Awemu, E.M. and Erhardt, G., Genetic structure and differentiation of 12 African Bos indicus and Bos taurus cattle breeds, inferred from protein and microsatellite polymorphisms, J. Anim. Breed. Genet., 2005, vol. 122, pp. 12–20.
Kumar, P., Freeman A.R., Loftus R.T., et al., Admixture analysis of South Asian cattle, Heredity, 2003, vol. 91, pp. 43–50.
Loftus, R.T., MacHugh, D.E., Bradley, D.G., et al., Evidence for two independent domestications of cattle, Proc. Natl. Acad. Sci. U.S.A., 1994, vol. 91, pp. 2757–2761.
Metta, M., Kanginakudru, S., Gudiseva, N., et al., Genetic characterization of the Indian cattle breeds, Ongole and Deoni (Bos indicus), using microsatellite markers-a preliminary study, BMC Genet., 2004, vol 5, p. 16.
Bicalho, H.M.S., Pimenta, C.G., Mendes, I.K.P., et al., Determination of ancestral proportions in synthetic bovine breeds using commonly employed microsatellite markers, Genet. Mol. Res., 2006, vol. 5, pp. 432–437.
Brenneman, R.A., Chase, C.C., Olson, T.A., et al., Genetic diversity among Angus, American Brahman, Senepol and Romosinuano cattle breeds, Anim. Genet., 2007, vol. 38, pp. 50–53.
Cervini, M., Henrique-Silva, F., Morati, N., et al., Genetic variability of 10 microsatellite markers in the characterization of Brazilian Nellore cattle (Bos indicus), Genet. Mol. Biol., 2006, vol. 29, pp. 486–490.
Egito, A.A., Paiva, S.R., Albuquerque, M.S., et al., Microsatellite based genetic diversity and relationships among ten Creole and commercial cattle breeds raised in Brazil, BMC Genet., 2007, vol. 8, p. 83.
Amos, W., Hoffman, J.I., Frodsham, A., et al., Automated binning of microsatellite alleles: problems and solutions, Mol. Ecol. Notes, 2006, vol. 7, pp. 10–14.
Raymond, M. and Rousset, F., An exact test for population differentiation, Evolution, 1995, vol. 49, pp.1280–1283.
Guo, S. and Thompson, E.A., Performing the exact test of Hardy-Weinberg proportion for multiple alleles, Biometrics, 1992, vol. 48, pp. 361–372.
Weir, B.S. and Cockerham, C.C., Estimating F statistics for the analysis of population structure, Evolution, 1984, vol. 38, pp. 1358–1370.
Marshall, T.C., Slate, J., Kruuk, L.E., et al., Statistical confidence for likelihood-based paternity inference in natural populations, Mol. Ecol., 1998, vol. 7, pp. 639–655.
Jamieson, A. and Taylor, S.C., Comparisons of three probability formulae for parentage exclusion, Anim. Genet., 1997, vol. 28, pp. 397–400.
Nei, M., Genetic distance between populations, Am. Nat., 1972, vol. 106, pp. 283–292.
Shriver, M.D., Jin, L., Boerwinkle, E., et al., A novel measure of genetic distance for highly polymorphic tandem repeat loci, Mol. Biol. Evol., 1995, vol. 12, pp. 914–920.
Page, R.D., TREEVIEW: an application to display phylogenetic trees on personal computers, Comput. Appl. Biosci., 1996, vol. 12, pp. 357–358.
Pritchard, J.K., Stephens, M., and Donnelly, P, Inference of population structure using multilocus genotype data, Genetics, 2000, vol. 155, pp. 945–959.
Falush, D., Stephens, M. and Pritchard, J.K., Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies, Genetics, 2003, vol. 164, pp. 1567–1587.
Liron, J.P., Ripoli, M.V., Garcia, P.P., Assignment of paternity in a judicial dispute between two neighbor Holstein dairy farmers, J. Forensic Sci., 2004, vol. 49, pp. 96–98.
Maudet, C., Luikart, G. and Taberlet, P., Genetic diversity and assignment tests among seven French cattle breeds based on microsatellite DNA analysis, J. Anim. Sci., 2002, vol. 80, pp. 942–950.
Martinez, R., Garcia, D., Gallego, J., et al., Genetic variability in Colombian Creole cattle populations estimated by pedigree information, J. Anim. Sci., 2008, vol. 86, pp. 545–552.
Cavalli-Sforza, L.L. and Edwards, A.W.F., Phylogenetic analysis: models and estimation procedures, Evolution, 1967, vol. 21, pp. 550–570.
Queller, D.C. and Goodnight, K.F., Estimating relatedness using genetic markers, Evolution, 1989, vol. 43, pp. 258–275.
Pamilo, P., Genotypic correlation and regression in social groups: multiple alleles, multiple loci and subdivided populations, Genetics, 1984, vol. 107, pp. 307–320.
Pamilo, P., Effect of inbreeding on genetic relatedness, Hereditas, 1985, vol. 103, pp. 195–200.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Gómez, Y.M., Fernández, M., Rivera, D. et al. Genetic characterization of Colombian Brahman cattle using microsatellites markers. Russ J Genet 49, 737–745 (2013). https://doi.org/10.1134/S1022795413070041
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795413070041