Abstract
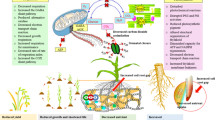
This paper reviews the most recent research progress in the field of salt tolerance for plants such as Arabidopsis, tomato, wheat, rice and cotton. Salinity tolerance is defined and classified, and research advances in the physiology, cellular biology and molecular biology of salt tolerance are presented. Additionally, transgenic breeding advances are profiled and the studies on quantitative trait locus are given; finally, an outlook for future salinity resistance research is proposed.
Similar content being viewed by others
References
Yu, S.W. and Tanag, Z.C., Plants Physiology and Molecular Biology, Beijing: Science Press, 1998.
Zhu, J.K., Plant Salt Tolerance, Trends Plant Sci., 2001, vol. 2, no. 6, pp. 66–71.
Cuartero, J., Bolarin, M.C., Asins, M.J., and Moreno, V., Increasing Salt Tolerance in Tomato, J. Exp. Bot., 2006, vol. 57, pp. 1045–1058.
Pan, R., Wang, X.J., and Li, N.H., Plant Physiology, Beijing: Higher Education Press, 2008.
Katerji, N., Hoorn, J.W., Hamdy, A., and Mastrorilli, M., Salinity Effect on Crop Development and Yield, Analysis of Salt Tolerance According to Several Classification Methods, Agric. Water Manage., 2003, vol. 62, pp. 37–66.
Chen, J.M., Research Advance in the Tolerance of Plant to Salt, Jiangsu, J. Agric. Sci., 2006, vol. 34, no. 14, pp. 248–254.
Jiang, Y., Lü, Y.J., and Zhu, S.J., Advance in Studies of the Mechanism of Salt Tolerance and Controlling of Salt Damage in Upland Cotton, Cott. Sci., 2006, vol. 18, no. 4, pp. 248–254.
Wang, W., Pan, Z.J., and Pan, Q.B., Studying Progress in Salt-Tolerant Characters of Crops, Acta Agric. Jiangxi, 2009, vol. 21, no. 2, pp. 30–33.
James, R.A., Munns, R., von Caemmere, S., et al., Photosynthetic Capacity is Related to the Cellular and Subcellular Partitioning of Na+, K+, and Cl− in Salt-Affected Barley and Durum Wheat, Plant Cell Environ., 2006, vol. 29, pp. 2185–2197.
Müller, I.S., Gilliham, M., Jha, D., and Mayo, G.M., Shoot Na+ Exclusion and Increased Salinity Tolerance Engineered by Cell Type-Specific Alteration of Na+ Transport in Arabidopsis, Plant Cell, 2009, vol. 21, pp. 2163–2178.
Tûrkana, I. and Demiral, T., Recent Developments in Understanding Salinity Tolerance, Environ. Exp. Bot., 2009, vol. 67, no. 1, pp. 2–9.
Hoekstra, F.A., Golovina, E.A., and Buitink, J., Mechanisms of Plant Desiccation Tolerance, Trends Plant Sci., 2001, vol. 6, pp. 431–443.
Sun, J.C. and Wang, X.C., Studies in Genetic Engineering on Salt Resistance in Rice, J. Ningxia Agric. For. Sci. Technol., 2007, vol. 4, pp. 40–42.
Turchetto-Zolet, A.C., Margis-Pinheiro, M., and Margis, R., The Evolution of Pyrroline-5-Carboxylate Synthase in Plants: A Key Enzyme in Proline Synthesis, Mol. Genet. Genomics, 2009, vol. 281, pp. 87–97.
Liu, F.H., Guo, Y., Gu, D.M., et al., Salt Tolerance of Transgenic Plants with BADH cDNA, Acta Gene Sin., 1997, vol. 27, no. 2, pp. 151–155.
Valentina, M., Theodoulou, F.L., Guy, K., et al., Coordinate Induction of Glutathione Biosynthesis and Glutathione Metabolizing Enzymes is Correlated with Salt Tolerance in Tomato, FEBS Lett., 2003, vol. 554, pp. 417–421.
Salekdeh, G.H., Siopongco, J., Wade, L.J., et al., A Proteomic Approach to Analyzing Drought- and Salt-Responsiveness in Rice, Field Crops Res., 2002, vol. 76, pp. 199–219.
Achard, P., Cheng, H., Grauwe, L.D., et al., Integration of Plant Responses to Environmentally Activated Phytohormonal Signals, Science, 2006, vol. 311, pp. 91–94.
Wang, H., Liang, X., Wanm, Q., et al., Ethylene and Nitric Oxide Are Involved in Maintaining Ion Homeostasis in Arabidopsis Callus under Salt Stress, Planta, 2009, vol. 230, pp. 293–307.
Yang, L., Zu, Y.G., and Tang, Z.H., Ethylene Improves Arabidopsis Salt Tolerance Mainly via Retaining K+ in Shoots and Roots rather than Decreasing Tissue Na+ Content, Environ. Exp. Bot., 2010, doi:10.1016/j.envexpbot.2010.08.006.
Xin, C.S., Dong, H.Z., Tang, W., and Wen, S.M., Physiological and Molecular Mechanisms of Salt Injury and Salt Tolerance in Cotton, Acta Gossypii Sin., 2005, vol. 17, no. 5, pp. 309–313.
Lin, H.X., Yanagihara, S., Zhuang, J.Y., et al., Identification of QTL for Salt Tolerance in Rice via Molecular Markers, Chin. J. Rice Sci., 1998, vol. 12, no. 2, pp. 72–78.
Lin, H.X., Zhu, M.Z., Yano, M., et al., QTLs for Na+ and K+ Uptake of the Shoots and Roots Controlling Rice Salt Tolerance, Theor. Appl. Genet., 2004, vol. 108, pp. 253–260.
Gao, J.P. and Lin, H.X., A Significant Progress of Salt Resistance in Rice—the Salt Resistant QTL SKC1, Chin. Bull. Life Sci., 2005, vol. 17, no. 6, pp. 563–565.
Wang, B., Lan, T., and Wu, W.R., Mapping of QTLs for Na+ Content in Rice Seedlings under Salt Stress, Chin. J. Rice Sci., 2007, vol. 21, no. 6, pp. 585–590.
Yao, M.Z., Wang, J.F., Chen, H.Y., et al., Inheritance and QTL Mapping of Salt Tolerance in Rice, Rice Sci., 2005, vol. 12, no. 1, pp. 25–32.
Wu, Y.R., Yi, K.K., and Zhu, J.M., Selection for Salt Tolerance of Tomato Population on the Analysis of Phenotype Parameter, J. Zhejiang Univ. Agric. Life Sci., 1999, vol. 25, no. 6, pp. 645–649.
Gao, X.L. and Xiao, Q.M., The NO3-N Contents of Causing Physiological Barriers of Tomato in Sunlight Greenhouse, Liaoning Agric. Sci., 1997, vol. 1, pp. 8–13.
Foolad, M.R., Recent Advances in Genetics of Salt Tolerance in Tomato, Plant Cell, Tissue Organ Cult., 2004, vol. 76, no. 2, pp. 101–119.
Dadshani, S.A.W., Weidner, A., Buck-Sorlin, G.H., et al., QTL Analysis for Salt Tolerance in Barley, Deutsoher Tropentag, 2004, pp. 5–7.
Zhu, Z.H., Hu, R.H., and Song, J.Z., Effects on the Seedlings of Different Kinds of Wheat with Salt Treatment, J. Nat. Resour., 1996, vol. 4, pp. 25–29.
Wu, Y.Q., Liu, L.X., and Guo, H.J., Mapping QTL for Salt Tolerant Traits in Wheat, J. Nucl. Agr. Sci., 2007, vol. 21, no. 6, pp. 545–549.
Ren, Z.H., Zheng, Z.M., Chinnusamy, V., et al., RAS1, a Quantitative Trait Locus for Salt Tolerance and ABA Sensitivity in Arabidopsis, Proc. Natl. Acad. Sci. U.S.A., 2010, vol. 107, no. 12, pp. 5669–5674.
Luo, Q.Y., Yu, B.J., Liu, Y.L., et al., The Mixed Inheritance Analysis of Salt Resistance in Cultivars of Glycine max, Soybean Sci., 2004, vol. 23, no. 4, pp. 239–244.
Shi, H.Z., Ishitani, M., Kim, C., and Zhu, J.K., The Arabidopsis thaliana Salt Tolerance Gene SOS1 Encodes a Putative Na+/H+ Antiporter, Proc. Natl. Acad. Sci. U.S.A., 2000, vol. 97, no. 12, pp. 6896–6901.
Liu, J.P., Ishitani, M., Halfter, U., et al., The Arabidopsis thaliana SOS2 Gene Encodes a Protein Kinase That Is Required for Salt Tolerance, Proc. Natl. Acad. Sci. U.S.A., 2000 vol. 97, no. 7, pp. 3730–3734.
Berthomieu, P., Conjro, G., Nublat, A., et al., Functional Analysis of AtHKT1 in Arabidopsis Shows That Na+ Recirculation by the Phloem Is Crucial for Salt Tolerance, EMBO J., 2003, vol. 22, no. 9, pp. 2004–2014.
Liu, Z.L., Huang, C.L., Zhang, X.H., and Wu, Z.Y., Application of Trehalose and the Study Progress of Trehalose Synthase Gene TPS in Transgenic Plants, Chin. Agric. Sci. Bull., 2009, vol. 25, no. 6, pp. 54–58.
Gaxiola, R.A., Li, J.S., Undurraga, S., et al., Drought- and Salt-Tolerant Plants Result from Over-Expression of the AVP1 H+-Pump, Proc. Natl. Acad. Sci. U.S.A., 2001, vol. 98, pp. 11444–11449.
Choi, W., Baek, D., Oh, D.H., et al., NKS1, Na+- and K+-Sensitive 1, Regulates Ion Homeostasis in an SOS — Independent Pathway in Arabidopsis, Phytochemistry, 2011, vol. 72, pp. 330–336.
Liu, C.F., Zou, J., and Chen, X.B., Advances in DREB Transcription Factors and Plant Abiotic Stress Tolerance, Biotechnol. Bull., 2010, vol. 10, pp. 26–30.
Lu, S.Y., Zhao, G.R., Wu, A.M., et al., Molecular Cloning of a Cotton Phosphatase Gene and Its Functional Characterization, Biokhimiya (Moscow), 2010, vol. 75, no. 1, pp. 85–94.
Wu, C.A., Yang, G.D., Meng, Q.W., and Zheng, C.C., The Cotton GhNHX1 Gene Encoding a Novel Putative Tonoplast Na+/H+ Antiporter Plays an Important Role in Salt Stress, Plant Cell Physiol., 2004, vol. 45, no. 5, pp. 600–607.
Fukuda, A., Nakamura, A., Tagiri, A., et al., Function, Intracellular Localization and the Importance in Salt Tolerance of a Vacuolar Na+/H+ Antiporter from Rice, Plant Cell Physiol., 2004, vol. 45, pp. 146–159.
Ren, Z.H., Gao, J.P., Li, L.G., et al., A Rice Quantitative Trait Locus for Salt Tolerance Encodes a Sodium Transporter, Nat. Genet., 2005, vol. 37, pp. 1141–1146.
Motohashi, T., Nagamiya, K.J., Prodhan, S.H., et al., Production of Salt Stress Tolerant Rice by Overexpression of the Catalase Gene, KatE, Derived from Escherichia coli AsPac, J. Mol. Biol. Biotechnol., 2010, vol. 18, no. 1, pp. 37–41.
Mukhopadhyay, M., Vij, S., and Tyagi, A.K., Overexpression of a Zinc-Finger Protein Gene from Rice Confers Tolerance to Cold, Dehydration, and Salt Stress in Transgenic Tobacco, Proc. Natl. Acad. Sci. U.S.A., 2004, vol. 101, no. 16, pp. 6309–6314.
Zhao, F.Y., Guo, S.L., Zhang, H., and Zhao, Y.X., Expression of Yeast SOD2 in Transgenic Rice Results in Increased Salt Tolerance, Plant Sci., 2006, vol. 170, pp. 216–224.
Shi, L., Gan, X.Y., Chen, Y.C., et al., Cloning and Sequence Analysis of Betaine Aldehyde Dehydrogenase Gene from Haloxylon ammodendron, Acta Bot. Bor.-Occid. Sin., 2010, vol. 30, no. 2, pp. 223–228.
Wegner, L.H. and Raschke, K., Ion Channels in the Xylem Parenchyma of Barley Roots—a Procedure to Isolate Protoplasts from This Tissue and a Patch-Clamp Exploration of Salt Passageways into Xylem Vessels, Plant Physiol., 1994, vol. 105, pp. 799–813.
Wang, J., Zuo, K., and Wu, W., Expression of a Novel Antiporter Gene from Brassica napus Resulted in Enhanced Salt Tolerance in Transgenic Tobacco Plants, Biol. Plantarum, 2004, vol. 48, pp. 509–515.
Zhang, Q.X., Xu, X.F., and Wang, Y., Isolation and Preliminary Function Analysis of a Na+/H+ Antiporter Gene from Malus zumi, Afr. J. Biotechnol., 2009, vol. 8, pp. 4774–4781.
Dong, Y.Z., Construction of Vector with IMT1 and Its Gene Expression in Transgenic Tobacco Leaf Cells Associated with Salt Tolerance, Acta Bot. Sin., 1999, vol. 41, no. 2, pp. 146–149.
Zong, Z.W. and Yang, X., Effect of mtlD Expression on Peanut Salt Tolerance, J. Anhui Agric. Sci., 2010, vol. 38, no. 20, pp. 10606–10607.
Takahashi, R., Liu, S.K., and Takano, T., Cloning and Functional Comparison of a High-Affinity K+ Transporter Gene Phahkt1 of Salt-Tolerant and Salt-Sensitive Reed Plants, J. Exp. Bot., 2007, vol. 58, nos. 15–16, pp. 4387–4395.
Yin, Y.L., Liang, J.S., and Liu, Q.Q., Cloning of HAL1 Gene from Saccharomyces cerevisiae and Construction of Its Plant Expression Vector, J. Yangzhou Univ. Agric. Life Sci., 2002, vol. 4, no. 23, pp. 27–29.
Shi, H.Z., Lee, B., Wu, S.J., and Zhu, J.K., Over-Expression of a Plasma Membrane Na+/H+ Antiporter Gene Improves Salt Tolerance in Arabidopsis thaliana, Nat. Biotechnol., 2003, vol. 21, pp. 81–85.
Wang, L.Y., Ding, G.H., and Li, L., Progress in Synthesis and Metabolism of Proline, J. Harbin Normal. Univ. Nat Sci., 2010, vol. 26, no. 2, pp. 84–89.
Cai, X.N., Yang, P., Fen, A.L., et al., Cloning of ThHKT1 Gene from Thellungiella halophile, Jiangsu J. Agric. Sci., 2006, vol. 6, pp. 21–24.
Duan, X.G., Yang, A.F., Gao, F., et al., Heterologous Expression of Vacuolar H+-PPase Enhances the Electrochemical Gradient Across the Vacuolar Membrane and Improves Tobacco Cell Salt Tolerance, Protoplasma, 2007, vol. 232, pp. 87–95.
Tang, R., Li, C., and Xu, K., Isolation, Functional Characterization and Expression Pattern of a Vacuolar Na+/H+ Antiporter Gene Trnhx1 from Trifolium repens L., Plant Mol. Biol. Rep., 2010, vol. 28, pp. 102–111.
Li, J.Y., Jiang, G.Q., Huang, P., et al., Over Expression of the Na+/H+ Antiporter Gene from Suaeda salsa Confers Cold and Salt Tolerance to Transgenic Arabidopsis thaliana, Plant Cell Tissue Organ Cult., 2007, vol. 90, pp. 41–48.
Koh, E.J., Song, W.Y., Lee, Y., et al., Expression of Yeast Cadmium Factor 1 (YCF1) Confers Salt Tolerance to Arabidopsis thaliana, Plant Sci., 2006, vol. 170, pp. 534–541.
Sheng, F.F., Yu, Y.J., and Yin, C.Q., Studies on Introducing Salt Resistance DNA of Dogbane into Cotton, Cott. Sci., 1995, vol. 7, no. 1, pp. 18–21.
Yu, Y.J., Variation of Characters in Upland Cotton (G. hirsutum) after Introduction by Exogenous DNA from Other Families, J. Shandong Agric. Univ. Nat. Sci., 1991, vol. 22, no. 4, pp. 335–340.
Li, N.Y. and Guo, Z.J., Over-Expression of Two Different Transcription Factors, OPBP1 and OsiWRKY, Enhances Resistance against Pathogen Attack and Salt Stress in Rice, Chin. J. Rice Sci., 2006, vol. 20, no. 1, pp. 13–18.
Martinez-Rodriguez, M.M., Estan, M.T., Moyano, E., et al., The Effectiveness of Grafting to Improve Salt Tolerance in Tomato When an ‘Excluder’ Genotype is Used as Scion. Environ. Exp. Bot., 2008, vol. 63, pp. 392–401.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
S. Yu and W. Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yu, S., Wang, W. & Wang, B. Recent progress of salinity tolerance research in plants. Russ J Genet 48, 497–505 (2012). https://doi.org/10.1134/S1022795412050225
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795412050225