Abstract
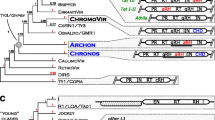
The rDNA locus of insects and other arthropods contains non-LTR retrotransposons (retroposons) that are specifically inserted into 28S rRNA genes. The most frequent retroposons are R1 and R2, but the mechanism of insertion and the functions of these mobile elements have not been studied in detail. A clone containing a full-length R1 retroposon copy was isolated from the cosmid library of Blattella germanica genes and sequenced. The amino acid sequences encoded by ORF1 of the R1 retroposon were subjected to bioinformatic analysis. It was found that ORF1 of this mobile element encodes a protein (ORF1p) belonging to the superfamily of zinc finger (CCHC) retroviral nucleocapsid proteins and contains two conserved RRM domains (RNA-recognizing motifs) identified on the basis of analysis of the secondary structure of this protein. The discovery of RRM domains in ORF1p of R1 retroposons can contribute to the understanding of the mechanisms of their retrotransposition. We revealed a coiled-coil motif in the N-terminal region of R1 ORF1p, which is similar to the coiled-coil domain involved in homo- or heteromultimerization of proteins and in protein-protein interactions. The domain organization of homologous Gag-like proteins of retroposons in some insects and fungi was found to be similar to the structure established for R1 ORF1p of B. germanica.
Similar content being viewed by others
References
Eickbush, T.H. and Jamburuthugoda, V.K., The Diversity of Retrotransposons and the Properties of Their Reverse Transcriptases, Virus Res., 2008, vol. 134, nos. 1–2, pp. 221–234.
Kapitonov, V.V. and Tempel, S.J.J., Simple and Fast Classification of Non-LTR Retrotransposons Based on Phylogeny of Their RT Domain Protein Sequences, Gene, 2009, vol. 448, no. 2, pp. 207–13.
Kojima, K.K. and Fujiwara, H., Cross-Genome Screening of Novel Sequence-Specific Non-LTR Retrotransposons: Various Multicopy RNA Genes and Satellites Are Selected as Targets, Mol. Biol. Evol., 2004, vol. 21, no. 2, pp. 207–217.
Kojima, K.K. and Fujiwara, H., Evolution of Target Specificity in R1 Clade Non-LTR Retrotransposons, Mol. Biol. Evol., 2003, vol. 20, no. 3, pp. 351–361.
Takahashi, H., Okazaki, S., and Fujiwara, H., A New Family of Site-Specific Retrotransposons, SART1, Is Inserted into Telomeric Repeats of the Silkworm, Bombyx mori, Nucl. Acids Res., 1997, vol. 25, no. 8, pp. 1578–1584.
Matsumoto, T., Hamada, M., Osanai, M., and Fujiwara, H., Essential Domains for Ribonucleoprotein Complex Formation Required for Retrotransposition of Telomere-Specific Non-Long Terminal Repeat Retrotransposon SART1, Mol. Cell. Biol., 2006, vol. 26, no. 13, pp. 5168–5179.
Khazina, E. and Weichenrieder, O., Non-LTR Retrotransposons Encode Noncanonical RRM Domains in Their First Open Reading Frame, Proc. Natl. Acad. Sci. USA, 2009, vol. 106, no. 3, pp. 731–736.
Heitkam, T. and Schmidt, T., BNR—a LINE Family from Beta vulgaris—Contains a RRM Domain in Open Reading Frame 1 and Defines a L1 Sub-Clade Present in Diverse Plant Genomes, Plant J., 2009, vol. 59, no. 6, pp. 872–882.
Maris, C., Dominguez, C., and Allain, F.H.T., The RNA Recognition Motif, a Plastic RNA-Binding Platform to Regulate Post-Transcriptional Gene Expression, FEBS J., 2005, vol. 272, no. 9, pp. 2118–2131.
Lunde, B.M., Moore, C., and Varani, G., RNA-Binding Proteins: Modular Design for Efficient Function, Nat. Rev. Mol. Cell Biol., 2007, vol. 8, no. 6, pp. 479–490.
Chen, Y. and Varani, G., Protein Families and RNA Recognition, FEBS J., 2005, vol. 272, no. 9, pp. 2088–2097.
Sding, J., Biegert, A., and Lupas, A.N., The HHpred Interactive Server for Protein Homology Detection and Structure Prediction, Nucl. Acids Res., 2005, vol. 33, pp. W244–W248.
Kagramanova, A.S., Kapelinskaya, T.V., Korolev, A.L., and Mukha, D.V., R1 and R2 Retrotransposons of German Cockroach Blatella germanica: A Comparative Study of 5’-Truncated Copies Integrated into the Genome, Mol. Biol. (Moscow), 2007, vol. 41, no. 4, pp. 546–553.
Tamura, K., Dudley, J., Nei, M., and Kumar, S., MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0, Mol. Biol. Evol., 2007, vol. 24, no. 8, pp. 1596–1599.
Zhang, Y., I-TASSER Server for Protein 3D Structure Prediction, BMC Bioinform., 2008, vol. 9, p. 40.
Inoue, K., Mizuno, T., Wada, K., and Hagiwara, M., Novel RING Finger Proteins, Air1p and Air2p, Interact with Hmt1p and Inhibit the Arginine Methylation of Npl3p, J. Biol. Chem., 2000, vol. 275, no. 42, pp. 32793–32799.
Sickmier, E.A., Frato, K.E., Shen, H., et al., Structural Basis for Polypyrimidine Tract Recognition by the Essential Pre-mRNA Splicing Factor U2AF65, Mol. Cell, 2006, vol. 23, no. 1, pp. 49–59.
Sawicka, K., Bushell, M., Spriggs, K.A., and Willis, A.E., Polypyrimidine-Tract-Binding Protein: A Multifunctional RNA-Binding Protein, Bioch. Soc. Trans., 2008, vol. 36, no. 4, pp. 641–647.
Allain, F.H., Bouvet, P., Dieckmann, T., and Feigon, J., Molecular Basis of Sequence-Specific Recognition of Pre-Ribosomal RNA by Nucleolin, EMBO J., 2000, vol. 19, no. 24, pp. 6870–6881.
Bae, E., Reiter, N.J., Bingman, C.A., et al., Structure and Interactions of the First Three RNA Recognition Motifs of Splicing Factor prp24, J. Mol. Biol., 2007, vol. 367, no. 5, pp. 1447–1458.
Bolognani, F., Contente-Cuomo, T., and Perrone-Bizzozero, N.I., Novel Recognition Motifs and Biological Functions of the RNA-Binding Protein HuD Revealed by Genome-Wide Identification of Its Targets, Nucleic Acids Res., 2010, vol. 38, no. 1, pp. 117–130.
Gibrat, J.F., Madej, T., and Bryant, S.H., Surprising Similarities in Structure Comparison, Curr. Opin. Struct. Biol., 1996, vol. 6, no. 3, pp. 377–385.
Martin, S.L., The ORF1 Protein Encoded by LINE-1: Structure and Function during L1 Retrotransposition, J. Biomed. Biotechnol., 2006, vol. 2006, pp. 45621–45626.
Dziembowski, A., Ventura, A-P., Rutz, B., et al., Proteomic Analysis Identifies a New Complex Required for Nuclear Pre-mRNA Retention and Splicing, EMBO J., 2004, vol. 23, no. 24, pp. 4847–4856.
Grigoryan, G. and Keating, A.E., Structural Specificity in Coiled-Coil Interactions, Curr. Opin. Struct. Biol., 2008, vol. 18, no. 4, pp. 477–483.
Casacuberta, E. and Pardue, M-L., HeT-A Elements in Drosophila virilis: Retrotransposon Telomeres Are Conserved across the Drosophila Genus, Proc. Natl. Acad. Sci. USA, 2003, vol. 100, no. 24, pp. 14091–14096.
Preker, P.J. and Keller, W., The HAT Helix, a Repetitive Motif Implicated in RNA Processing, Trends Biochem. Sci., 1998, vol. 23, no. 1, pp. 15–16.
Blatch, G.L. and Lassle, M., The Tetratricopeptide Repeat: A Structural Motif Mediating Protein-Protein Interactions, BioEssays, 1999, vol. 21, no. 11, pp. 932–939.
Kobe, B. and Deisenhofer, J., The Leucine-Rich Repeat: A Versatile Binding Motif, Trends Biochem. Sci., 1994, vol. 19, no. 10, pp. 415–421.
Lee, C., Hong, B.S., Choi, J.M., et al., Structural Basis for Inhibition of the Replication Licensing Factor Cdt1 by Geminin, Nature, 2004, vol. 430, no. 7002, pp. 913–917.
Maiorano, D., Rul, W., and Mechali, M., Cell Cycle Regulation of the Licensing Activity of Cdt1 in Xenopus laevis, Exp. Cell Res., 2004, vol. 295, no. 1, pp. 138–149.
Nefedova, L.N. and Kim, A.I., Molecular Evolution of Mobile Elements of the gypsy Group: A Homolog of the gag Gene in Drosophila, Russ. J. Genet., 2009, vol. 45, no. 1, pp. 23–29.
Kroutter, E.N., Belancio, V.P., Wagstaff, B.J., and RoyEngel, A.M., The RNA Polymerase Dictates ORF1 Requirement and Timing of LINE and SINE Retrotransposition, PLoS Genet., 2009, vol. 5, no. 4, p. e1000458.
Rashkova, S., Athanasiadis, A., and Pardue, M.-L., Intracellular Targeting of Gag Proteins of the Drosophila Telomeric Retrotransposons, J. Virol., 2003, vol. 77, no. 11, pp. 6376–6384.
Zimin, A.V., Smith, D.R., Sutton, G., and Yorke, J.A., Assembly Reconciliation, Bioinformatics, 2008, vol. 24, no. 1, pp. 42–45.
Clark A.G., Eisen M.B., et al. Evolution of Genes and Genomes on the Drosophila Phylogeny, Nature, 2007, vol. 450, no. 7167, pp. 203–218.
Villasante, A., Abad, J.P., Planell, R., et al., Drosophila Telomeric Retrotransposons Derived from an Ancestral Element That Was Recruited to Replace Telomerase, Genome Res., 2007, vol. 17, no. 12, pp. 1909–1918.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © T.V. Kapelinskaya, A.S. Kagramanova, A.L. Korolev, D.V. Mukha, 2011, published in Genetika, 2011, Vol. 47, No. 2, pp. 149–158.
Rights and permissions
About this article
Cite this article
Kapelinskaya, T.V., Kagramanova, A.S., Korolev, A.L. et al. First open reading frame protein (ORF1p) of the Blattella germanica R1 retroposon and phylogenetically close GAG-like proteins of insects and fungi contain RRM domains. Russ J Genet 47, 129–138 (2011). https://doi.org/10.1134/S1022795410121038
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795410121038