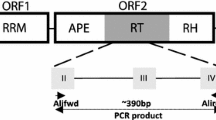
Abstract
Using cosmid vector, a gene library of German cockroach Blattella germanica was constructed. From this library, clones containing full-length copies of two subfamilies of R1 retroposons were selected. Retroposons R1 of German cockroach belonging to different subfamilies were shown to be different in domain organization of the ORF2 C-terminal region. For the first time, retroposons transmembrane domains were identified in the sequences of R1. It was demonstrated that two retroposon R1 subfamilies of German cockroach arose as a result of intragenomic divergence rather than via horizontal transfer of alien mobile element into cockroach genome. The differences in domain organization appeared not as a result of saltatory recombination processes, but as a consequence of gradual mutation accumulation, which led to either degeneration, or to domain formation.
Similar content being viewed by others
References
Finnegan, D.J., Transposable Elements: How Non-LTR Retrotransposons Do It, Current Biol., 1997, vol. 7, pp. 245–248.
Eickbush, T.H. and Jamburuthugoda, V.K., The Diversity of Retrotransposons and the Properties of Their Reverse Transcriptases, Virus Res., 2008, vol. 134, pp. 221–234.
Malik, H.S., Burke, W.D., and Eickbush, T.H., The Age and Evolution of Non-LTR Retrotransposable Elements, Mol. Biol. Evol., 1999, vol. 16, pp. 793–805.
Kojima, K.K. and Fujiwara, H., Evolution of Target Specificity in R1 Clade Non-LTR Retrotransposons, Mol. Biol. Evol., 2003, vol. 20, pp. 351–361.
Perez-Gonzalez, C.E. and Eickbush, T.H., Dynamics of R1 and R2 Elements in the rDNA Locus of Drosophila simulans, Genetics, 2001, vol. 158, pp. 1557–1567.
Gentile, K.L., Burke, W.D., and Eickbush, T.H., Multiple Lineages of R1 Retrotransposable Elements Can Coexist in the rDNA Loci of Drosophila, Mol. Biol. Evol., 2001, vol. 18, pp. 235–245.
Burke, W.D., Eickbush, D.G., Xiong, Y., et al., Sequence Relationship of Retrotransposable Elements R1 and R2 within and between Divergent Insect Species, Mol. Biol. Evol., 1993, vol. 10, pp. 163–185.
McGraw, J.E. and Brookfield, J.F., The Interaction between Mobile DNAs and Their Hosts in a Fluctuating Environment, J. Theor. Biol., 2006, vol. 243, pp. 13–23.
Kagramanova, A.S., Kapelinskaya, T.V., Korolev, A.L., and Mukha, D.V., R1 and R2 Retrotransposons of German Cockroach Blatella germanica: A Comparative Study of 5’-Truncated Copies Integrated into the Genome, Mol. Biol., (Moscow), 2007, vol. 41, pp. 546–553.
Kumar, S., Dudley, J., Nei, M., and Tamura, K., MEGA: A Biologist-Centric Software for Evolutionary Analysis of DNA and Protein Sequences, Briefings Bioinf., 2008, vol. 9, pp. 299–306.
Letunic, I., Doerks, T., and Bork, P., SMART 6: Recent Updates and New Developments, Nucleic Acids Res., 2008, vol. 37, pp. D229–D232.
Nielsen, M., Lundegaard, C., Lund, O., and Petersen, T.N., CpHModels-3.0: Remote Homology Modeling Using Structure Guided Profile Sequence Alignment and Double-Sided Baseline Corrected Scoring Scheme, Worning Abstract at the CASP8 Conference 193.
Aires, J.R., Köhler, T., Nikaido, H., and Plesiat, P., Involvement of an Active Efflux System in the Natural Resistance of Pseudomonas aeruginosa to Aminoglycosides, Antimicrob. Agents Chemother., 1999, vol. 43, pp. 2624–2628.
Wong, K.K.Y., Brinkman, F.S.L., Benz, R.S., and Hancock, R.E.W., Evaluation of a Structural Model of Pseudomonas aeruginosa Outer Membrane Protein OprM, an Efflux Component Involved in Intrinsic Antibiotic Resistance, J. Bacteriol., 2001, vol. 183, pp. 367–374.
Malik, H.S. and Eickbush, T.H., NeSL-1, an Ancient Lineage of Site Specific Non-LTR Retrotransposones from Caenorhabditis elegans, Genetics, 2000, vol. 154, pp. 193–203.
Ohno, S., Evolution by Gene Duplication, New York: Springer, 1970.
Deredec, A., Burt, A., and Godfray, H.C.J., The Population Genetics of Using Homing Endonuclease Genes in Vector and Pest Management, Genetics, 2008, vol. 179, pp. 2013–2026.
Glenner, H., Thomsen, P.F., Hebsgaard, M.B., et al., The Origin of Insects, Science, 2006, vol. 314, pp. 1883–1884.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A.S. Kagramanova, T.V. Kapelinskaya, A.L. Korolev, D.V. Mukha, 2010, published in Genetika, 2010, Vol. 46, No. 8, pp. 1041–1049.
Rights and permissions
About this article
Cite this article
Kagramanova, A.S., Kapelinskaya, T.V., Korolev, A.L. et al. Domain organization of the ORF2 C-terminal region of the German cockroach retroposon R1. Russ J Genet 46, 924–931 (2010). https://doi.org/10.1134/S102279541008003X
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102279541008003X