Abstract
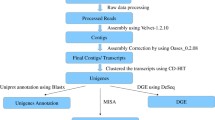
The roots of Panax ginseng C.A. Meyer, known as Korean ginseng have been a valuable and important folk medicine in East Asian countries. It mainly used to maintain the homeostasis of the human body, with the presence of ginsenosides and non-saponin compounds like phenol compounds, acidic polysaccharides and polyethylene compounds. Functional genomics aid to annotate EST sequences based on gene ontology. In this study, we focused, genes which involve in secondary metabolic pathways and to visualize temporal changes of gene expression in ginseng hairy roots with methyl ester methyl jasmonate (MeJA) along with non-treated hairy roots. A 5.774 EST clones were clustered and assembled into 501 contigs and 2.955 singletons. Annotations categorized with molecular functions, biological processes, cellular compounds of gene ontological terms and biochemical functions, enzyme commission number, and metabolic pathways are assigned through Kyoto Encyclopedia of Genes and Genomes database. Comparatively, EST sequences are assigned to cellular process, metabolic process, biotic and abiotic stress stimuli, developmental and biological regulations and transports are up-regulated 2–3 fold in MeJA treated hairy roots. 46 different sub groups of enzymes found in the MeJA treated plants. These annotated ESTs represents a significant proportion of the P. ginseng and provides molecular resource for develop microarray to study genes expressions to development, metabolism and reproduction.
Similar content being viewed by others
References
Vogler, B.K., Pittler, M.H., and Ernst, E., The Efficacy of Ginseng, a Systematic Review of Randomized Clinical Trials, Eur. J. Clin. Pharmacol., 1999, vol. 55, pp. 567–575.
Sticher, O., Getting to the Root of Ginseng, Chemtech, 1998, vol. 28, pp. 26–32.
Kwang-tae, Choi., Botanical Characteristics, Pharmacological Effects and Medicinal Components of Korean Panax ginseng C.A. Meyer, Acta Pharmacologica Sinica, 2008, vol. 29, no. 9, pp. 1109–1118.
Haralampidis, K., Trojanowska, M., and Osbourn, A.E., Biosynthesis of Triterpenoid Saponins in Plants, Adv. Biochem. Eng. Biotechnol., 2002, vol. 75, pp. 31–49.
Wasternack, C. and Parthier, B., Jasmonate-Signaled Plant Gene Expression, Trends Plant Sci., 1997, vol. 2, pp. 1360–1385.
Beckers, G.J.M. and Spoel, S.H., Fine Tuning Plant Defense Signaling: Salicylate versus Jasmonate, Plant Biol., 2008, vol. 8, pp. 1–10.
Yukimine, Y., Tabata, H., Higashi, Y., and Hara, Y., Methyl Jasmonate-Induced Overproduction of Paclitaxel and Baccation III in Taxus Cell Suspension Cultures, Nat. Biotechnol., 1996, vol. 14, pp. 1129–1132.
Martin, D., Throll, D., Gershenzon, J., and Bohlmann, J., Methyl Jasmonate Induces Traumatic Resin Ducts, Terpenoid Resion Biosynthesis, and Terpenoid Accumulation in Developing Xylem of Norway Spruce Stem, Plant Physiol., 2002, vol. 129, pp. 1002–1028.
Choi, D., Jung, J.D., Ha, Y.I., et al., Analysis of Transcripts in Methyl Jasmonate-Treated Ginseng Hairy Roots to Identify Genes Involved in the Biosynthesis of Ginsenosides and Other Secondary Metabolites, Plant Cell Rep., 2005, vol. 23, pp. 557–566.
Conesa, A. and Götz, S., Blast2GO: a Comprehensive Suite for Functional Analysis in Plant Genomics, Int. J. Plant Genomics, 2008, vol. 28, pp. 619–632.
Conesa, A., Götz, S., García-Gómez, J.M., et al., Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research, Bioinformatics, 2005, vol. 21, pp. 3674–3676.
Götz, S., García-Gómez, J.M., Terol, J., et al., High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite, Nucleic Acids Res., 2008, vol. 36, no. 10, pp. 3420–3435.
Pal, D., On Gene Ontology and Function Annotation, Bioinformation, 2006, vol. 1, no. 3, pp. 97–98.
Gene Ontology Consortium: The Gene Ontology (GO) Project in 2006, Nucleic Acids Res., 2006, vol. 34(Data-base issue), pp. D322–D326.
Dauchot, N., Mingeot, N., and Purnelle, B., Construction of 12 EST Libraries and Characterization of a 12,226 EST Dataset for Chicory (Cichorium intybus) Root, Leaves and Nodules in the Context of Carbohydrate Metabolism Investigation, BMC Plant Biol., 2009, vol. 9, p. 14.
Kanehisa, M. and Goto, S., KEGG: Kyoto Encyclopaedia of Genes and Genomes Nucleic Acids Res., 2000, vol. 28, pp. 27–30.
Morris, P.C., Kumar, A., Bowles, D.J., and Cuming, A.C., Osmotic Stress and Abscisic Acid Regulate the Expression of the Em Gene of Wheat, Eur. J. Biochem., 1990, vol. 190, pp. 625–630.
Masoudi-Nejad, A., Tonomura, K., Kawashima, S., et al., EGassembler: Online Bioinformatics Service for Large-Scale Processing, Clustering and Assembling ESTs and Genomic DNA Fragments, Nuclic Acids Res., 2006, vol. 34, pp. W459–W462.
Huang, X. and Madan, A., CAP3: A DNA Sequence Assembly Program, Genome Res., 1999, vol. 9. pp. 868–877.
Quevillon, E., Silventoinen, V., Pillai, S., et al., Inter-ProScan: Protein Domains Identifier, Nuclic Acids Res., 2005, vol. 33, pp. W116–W120.
Botton, A., Galla, G., Conesa, A., et al., Large-Scale Gene Ontology Analysis of Plant Transcriptome-Derived Sequences Retrieved by AFLP Technology, BMC Genomics, 2008, vol. 9, p. 347.
Adhikari, B.N., Wall, D.H., and Adams, B.J., Desiccation Survival in an Antarctic Nematode: Molecular Analysis Using Expressed Sequenced Tags, BMC Genomics, 2009, vol. 10, p. 69.
Zhaoa, W.J., Zhanga, H., Boa, X., et al., Generation and Analysis of Expressed Sequence Tags from a cDNA Library of Moniezia expansa, Mol. Biochem. Parasitol., 2009, vol. 164, pp. 80–85.
Rischer, H., Orešič, M., Seppänen-Laakso, T., et al., Gene-to-Metabolite Networks for Terpenoid Indole Alkaloid Biosynthesis in Catharanthus roseus Cells, Proc. Natl. Acad. Sci. USA, 2006, vol. 103, no. 14, pp. 5614–5619.
Purev, M., Kim, M.K., Samdan, N., and Yang, D.C., Isolation of a Novel Fructose-1,6-Bisphosphate Aldolase Gene from Codonopsis lanceolata and Analysis of the Response of This Gene to Abiotic Stresses, Mol. Biol. (Moscow), 2008, vol. 42, no. 2, pp. 206–213.
Reed, J.L., Famili, I., Thiele, I., and Palsson, B.O., Towards Multidimensional Genome Annotation, Nat. Rev. Genet., 2006, vol. 7, no. 2, pp. 130–141.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © S. Sathiyamoorthy, J.-G. In, S. Gayathri, Y.Ju Kim, D.-Ch. Yang, 2010, published in Genetika, 2010, Vol. 46, No. 7, pp. 932–939.
The article is published in the original.
Rights and permissions
About this article
Cite this article
Sathiyamoorthy, S., In, J.G., Gayathri, S. et al. Gene ontology study of methyl jasmonate-treated and non-treated hairy roots of Panax ginseng to identify genes involved in secondary metabolic pathway. Russ J Genet 46, 828–835 (2010). https://doi.org/10.1134/S1022795410070070
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795410070070