Abstract
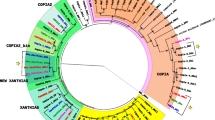
Retrotransposons of the gypsy group of Drosophila melanogaster that are structurally similar to retroviruses of vertebrates occupy an important place among retroelements of eukaryotes. The infectious abilities of some retrotransposons of this group (gypsy, ZAM, and Idefix) have been demonstrated experimentally, and therefore they are true retroviruses. It is supposed that retrotransposons can evolve acquiring new components, the sources of which remain to be elucidated. In this work, the CG4680gene (Gag related protein, Grp) homologous to gag of retrotransposons of the gypsy group has been identified in the genome of D. melanogaster and characterized. The Grp gene product has a highly conserved structure in different species of the Drosophilidae family and is under of purifying selection, which suggests its important genomic function in Drosophila. In view of the earlier data, it can be concluded that homologous genes of all components of gypsy retrotransposons are present in the Drosophila genome. These genes can be both precursors and products of domestication of retrovirus genes.
Similar content being viewed by others
References
Hardman, N., Structure and Function of Repetitive DNA in Eukaryotes, Biochem. J., 1986, vol. 234, no. 1, pp. 1–11.
Temin, H.M., Origin of Retroviruses from Cellular Moveable Genetic Elements, Cell, 1980, vol. 21, no. 3, pp. 599–600.
Temin, H.M., Reverse Transcriptases: Retrons in Bacteria, Nature, 1989, vol. 339, no. 6222, pp. 254–255.
Song, S.U., Gerasimova, T., Kurkulos, M., et al., An env-Like Protein Encoded by a Drosophila Retroelement: Evidence That gypsy Is an Infectious Retrovirus, Genes Dev., 1994, vol. 8, no. 17, pp. 2046–2057.
Kim, A.I., Lyubomirskaya, N.V., Belyaeva, E.S., et al., The Introduction of Transposionally Active Copy of a Retrotransposon gypsy into the Stable Strain of Drosophila melanogaster, Mol. Gen. Genet., 1994, vol. 242, no. 4, pp. 472–477.
Leblanc, P., Desset, S., Giorgi, F., et al., Life Cycle of an Endogenous Retrovirus, ZAM, in Drosophila melanogaster, J. Virol., 2000, vol. 74, no. 22, pp. 10658–10669.
Boeke, J.D, Eickbush, T.H, Sandmeyer, S.B, and Voytas, D.F, Index of Viruses—Metaviridae, in ICTVdB—The Universal Virus Database, Version 4, Buchen-Osmond, C., Ed., New York: Columbia Univ., 2006.
Lung, O. and Blissard, G.W., A Cellular Drosophila melanogaster Protein with Similarity to Baculovirus F Envelope Fusion Proteins, J. Virol., 2005, vol. 79, no. 13, pp. 7979–7989.
Malik, H.S. and Henikoff, S., Positive Selection of Iris, a Retroviral Envelope-Derived Host Gene in Drosophila melanogaster, PLoS Genet., 2005, vol. 1, no. 4, p. e44.
Nefedova, L.N. and Kim, A.I., Evolution from Retrotransposons to Retroviruses: The Source and Origin of env Gene, Zh. Obshch. Biol., 2007, vol. 68, no. 6, pp. 459–467.
Bowen, N.J. and McDonald, J.F., Drosophila Euchromatic LTR Retrotransposons Are Much Younger Than the Host Species in Which They Reside, Genome Res., 2001, vol. 11, no. 9, pp. 1527–1540.
Capy, P., Classification and Nomenclature of Retrotransposable Elements, Cytogenet. Genome Res., 2005, vol. 110, nos. 1–4, pp. 457–461.
McClure, M.A., Evolution of Retroposons by Acquisition or Deletion of Retrovirus-Like Genes, Mol. Biol. Evol., 1991, vol. 8, no. 6, pp. 835–856.
Mugnier, N., Biemont, C., and Vieira, C., New Regulatory Regions of Drosophila 412 Retrotransposable Element Generated by Recombination, Mol. Biol. Evol., 2005, vol. 22, no. 3, pp. 747–757.
Malik, H.S., Henikoff, S., and Eickbush, T.H., Poised for Contagion: Evolutionary Origins of the Infectious Abilities of Invertebrate Retroviruses, Genome Res., 2000, vol. 10, no. 9, pp. 1307–1318.
Doolittle, R.F., Feng, D.F., Johnson, M.S., and McClure, M.A., Origins and Evolutionary Relationships of Retroviruses, Q. Rev. Biol., 1989, vol. 64, no. 1, pp. 1–30.
Xiong, Y. and Eickbush, T.H., Origin and Evolution of Retroelements Based upon Their Reverse Transcriptase Sequences, EMBO J., 1990, vol. 9, no. 10, pp. 3353–3362.
Wan, H. and Wootton, J.C., A Global Compositional Complexity Measure for Biological Sequences: AT-Rich and GC-Rich Genomes Encode Less Complex Proteins, Comput. Chem., 2000, vol. 24, no. 1, pp. 71–94.
Rein, A., Henderson, L.E., and Levin, J.G., Nucleic-Acid-Chaperone Activity of Retroviral Nucleocapsid Proteins: Significance for Viral Replication, Trends Biochem. Sci., 1998, vol. 23, no. 8, pp. 297–301.
Gabus, C., Ivanyi-Nagy, R., Depollier, J., et al., Characterization of a Nucleocapsid-Like Region and of Two Distinct Primer tRNALys, 2 Binding Sites in the Endogenous Retrovirus Gypsy, Nucleic Acids Res., 2006, vol. 34, no. 20, pp. 5764–5777.
Nethe, M., Berkhout, B., and van der Kuyl, A.C., Retroviral Superinfection Resistance, Retrovirology, 2005, vol. 2, p. 52.
Dupressoir, A., Marceau, G., Vernochet, C., et al., Syncytin-A and syncytin-B, Two Fusogenic Placenta-Specific Murine Envelope Genes of Retroviral Origin Conserved in Muridae, Proc. Natl. Acad. Sci. USA, 2005, vol. 102, no. 3, pp. 725–730.
Brandt, J., Veith, A.M., and Volff, J.N., A Family of Neofunctionalized Ty3/gypsy Retrotransposon Genes in Mammalian Genomes, Cytogenet. Genome Res., 2005, vol. 110, nos. 1–4, pp. 307–317.
Campillos, M., Doerks, T., Shah, P.K., and Bork, P., Computational Characterization of Multiple Gag-Like Human Proteins, Trends Genet., 2006, vol. 22, no. 11, pp. 585–589.
Nefedova, L.N. and Kim, A.I., Evolution of Errantiviruses of Drosophila melanogaster, Strategy 2: From Retroviruses to Retrotransposons, Russ. J. Genet., 2007, vol. 43, no. 10, pp. 1388–1395.
Kaminker, J.S., Bergman, C.M., Kronmiller, B., et al., The Transposable Elements of the Drosophila melanogaster Euchromatin: A Genomics Perspective, Genome Biol., 2002, vol. 3, no. 12, RESEARCH0084.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © L.N. Nefedova, A.I. Kim, 2009, published in Genetika, 2009, Vol. 45, No. 1, pp. 30–37.
Rights and permissions
About this article
Cite this article
Nefedova, L.N., Kim, A.I. Molecular evolution of mobile elements of the gypsy group: A homolog of the gag gene in Drosophila . Russ J Genet 45, 23–29 (2009). https://doi.org/10.1134/S1022795409010037
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795409010037