Abstract
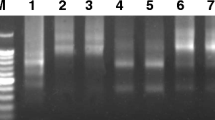
C banding, Ag-NOR staining, FISH with pTa71 (45S rDNA) and pTa794 (5S rDNA), and RAPD-PCR analysis were used to study the genome and chromosome polymorphism in four varieties (Frisson, Sparkle, Rondo, and Finale) and two genetic lines (Sprint-2 and SGE) of pea Pisum sativum L. A comparison of the C-banding patterns did not reveal any polymorphism within the varieties. The most significant between-variety differences were observed for the size of C bands on satellite chromosomes 4 and 7. All grain pea varieties (Frisson, Sparkle, and Rondo) had a large C band in the satellite of chromosome 4 and a medium C band in the region adjacent to the satellite thread on chromosome 7. C bands were almost of the same size in the genetic lines and vegetable variety Finale. In all accessions, 45S rDNA mapped to the secondary constriction regions of chromosomes 1, 3, and 5. The signal from chromosome 5 in the lines was more intense than in the varieties. Ag-NOR staining showed that the transcriptional activity of the 45S rRNA genes on chromosome 7 was higher than on chromosome 4 in all accessions. No more than four Ag-NOR-positive nucleoli were observed in interphase nuclei. Statistical analysis of the total area of Ag-NOR-stained nucleoli did not detect any significant difference between the accessions examined. RAPD-PCR analysis revealed high between-variety and low within-variety genomic polymorphism. Chromosomal and molecular markers proved to be promising for genome identification in pea varieties and lines.
Similar content being viewed by others
References
Borisov, A.Yu., Provorov, N.A., Tikhonovich, I.A., and Tsyganov, V.E., Genetic Control of the Interaction between Legumes and Nodule Bacteria, Genetika simbioticheskoi azotfiksatsii s osnovami selektsii (Genetics of Symbiotic Nitrogen Fixation and the Principles of Breeding), Tikhonovich, I. and Provorov, N.A., Eds., St. Petersburg: Nauka, 1998.
Shtark, O.Yu., Danilova, T.N., and Naumkina, T.S., Analysis of Pea (Pisum sativum L.) Initial Material for Selection of Varieties with a High Symbiotic Potential and the Choice of Parameters for Its Evaluation, Ekolog. Genet., 2006, vol. 4, no. 2, pp. 22–28.
Greilhuber, J. and Ebert, I., Genome Size Variation in Pisum sativum, Genome, 1994, vol. 37, no. 4, pp. 646–655.
Lamprecht, H., The Variation of Linkage and the Course of Crossingover, Agric. Hort. Genet., 1948, vol. 6, pp. 10–49.
Blixt, S., Cytology of Pisum: II. The Normal Karyotype, Agric. Hort. Genet., 1958, vol. 16, pp. 221–237.
Blixt, S., Cytology of Pisum: III. Investigation of Five Interchangel Lines and Coordination of Linkage Groups with Chromosomes, Agric. Hort. Genet., 1959, vol. 17, pp. 47–75.
Folkeson, D., Assignment of Linkage Segments to the Satellite Chromosomes 4 and 7 in Pisum sativum, Hereditas, 1990, vol. 112, pp. 257–263.
Folkeson, D., Assignment of Linkage Segments to Chromosomes 3 and 5 in Pisum sativum, Hereditas, 1990, vol. 112, pp. 249–256.
Hall, K.J., Parker, J.D., and Ellis, T.H.N., The Relationship between Genetic and Cytogenetic Maps of Pea: I. Standard and Translocation Karyotypes, Genome, 1997, vol. 40, pp. 744–754.
Hall, K.J., Parker, J.S., Ellis, T.H.N., et al., The Relationship between Genetic and Cytogenetic Maps of Pea: II. Physical Maps of Linkage Mapping Populations, Genome, 1997, vol. 40, pp. 755–769.
Rawlins, D.J., Highert, M.I., and Shaw, P.J., Localization of Telomeres in Plant Interphase Nuclei by in situ Hybridization and 3D Confocal Microscopy, Chromosoma, 1991, vol. 100, pp. 424–431.
Marchenko, A.N. and Gostimskii, S.A., Identification of a New Translocation in Pea, Dokl. Akad. Nauk SSSR, 1997, vol. 357, pp. 713–716.
Neumann, P., Nouzova, M., and Macas, J., Molecular and Cytogenetic Analysis of Repetitive DNA in Pea (Pisum sativum L.), Genome, 2001, vol. 44, pp. 716–728.
Neumann, P., Pozarkova, D., Vrana, J., et al., Chromosome Sorting and PCR-Based Physical Mapping in Pea (Pisum sativum L.), Chromosome Res., 2002, vol. 10, pp. 63–71.
Samatadze, T.E., Muravenko, O.V., Zelenin, A.V., and Gostimskii, S.A., Identification of Pea (Pisum sativum L.) Chromosomes Using C-Banding Analysis, Dokl. Akad. Nauk, 2002, vol. 387, no. 5, pp. 714–717.
Samatadze, T.E., Muravenko, O.V., Bolsheva, N.L., et al., Investigation of Chromosomes in Varieties and Translocation Lines of Pea Pisum sativum L. by FISH, Ag-NOR, and Differential DARI Staining, Russ. J. Genet., 2005, vol. 41, no. 12, pp. 1381–1388.
Lu, J., Knox, M.R., Ambrose, M.J., et al., Comparative Analysis of Genetic Diversity in Pea Assessed by RFLPand PCR-Based Methods, Theor. Appl. Genet., 1996, vol. 93, pp. 1103–1111.
Laucou, V., Haurogne, K., Ellis, N., and Rameau, C., Genetic Mapping in Pea: 1. RAPD-Based Genetic Linkage Map of Pisum sativum, Theor. Appl. Genet., 1998, vol. 97, pp. 905–915.
Rameau, C., Denoue, D., Fraval, F., et al., Genetic Mapping in Pea: 2. Identification of RAPD and SCAR Markers Linked to Genes Affecting Plant Architecture, Theor. Appl. Genet., 1998, vol. 97, pp. 916–928.
von Stackelberg, M., Linemann, S., Menke, M., et al., Identification of AFLP and STS Markers Closely Linked to the def Locus in Pea, Theor. Appl. Genet., 2003, vol. 106, pp. 1293–1299.
Janilla, P. and Sharma, B., RAPD and SCAR Markers for Powdery Mildew Resistance Gene er in Pea, Plant Breed., 2004, vol. 123, no. 3, pp. 271–274.
Loridon, K., McPhee, K., Morin, J., et al., Microsatellite Marker Polymorphism and Mapping in Pea (Pisum sativum L.), Theor. Appl. Genet., 2005, vol. 111, pp. 1022–1031.
Gostimskii, S.A., Kokaeva, Z.G., and Konovalov, F.A., Studying Plant Genome Variation Using Molecular Markers, Russ. J. Genet., 2005, vol. 41, no. 4, pp. 378–388.
Duc, G. and Messager, A., Mutagenesis of Pea (Pisum sativum L.) and the Isolation of Mutants for Nodulation and Nitrogen Fixation, Plant Sci., 1989, vol. 60, pp. 207–213.
Kneen, B.E. and LaRue, T.A., Nodulation Resistant Mutant of Pisum sativum L., J. Heredity, 1984, vol. 75, pp. 238–240.
Feenstra, W.J. and Jacobsen, E., Isolation of a Nitrate Reductase Deficient Mutant of Pisum sativum by Means of Selection for Chlorate Resistance, Theor. Appl. Genet., 1980, vol. 58, pp. 39–42.
Engvild, K.J., Nodulation and Nitrogen Fixation Mutants of Pea (Pisum sativum), Theor. Appl. Genet., 1987, vol. 74, pp. 711–713.
Kosterin, O.E. and Rozov, S.M., Mapping of the New Mutation blb and the Problem of Integrity of Linkage Group I, Pisum Genet., 1993, vol. 25, pp. 27–31.
Berdnikov, V.A., Rozov, S.M., and Bogdanova, V.S., Production of Pea Laboratory Line Series, in Chastnaya genetika rastenii (Specific Plant Genetics), Proc. Conf., Kievx, 1989, vol. 2, pp. 47–51.
Zelenina, D.A., Khrustaleva, A.M., and Volkov, A.A., Comparative Study of the Population Structure and Population Assignment of Sockeye Salmon Oncorhynchus nerka from West Kamshatka Based on RAPD-PCR and Microsatellite Polymorphism, Russ. J. Genet., 2006, vol. 42, no. 5, pp. 563–572.
Muravenko, O.V., Yurkevich, O.Yu., Bolsheva, N.L., et al., Comparison of Genomes of Eight Species of Sections Linum and Adenolinum from the Genus Linum Based on Chromosome Banding, Molecular Markers, and RAPD Analysis, Russ. J. Genet., 2008, (in press).
Fuchs, J., Kuhne, M., and Schubert, I., Assignment of Linkage Groups to Pea Chromosomes after Karyotyping and Gene Mapping by Fluorescent in situ Hybridization, Chromosoma, 1998, vol. 107, pp. 272–276.
Muravenko, O.V., Samatadze, T.E., Popov, K.V., et al., Polymorphism of Heterochromatic Regions of Flax Chromosomes, Biol. Membr., 1998, vol. 15, no. 6, pp. 670–678.
Muravenko, O.V., Amosova, A.V, Samatadze, T.E., et al., Chromosome Localization 5S and 45S Ribosomal DNA in the Genomes of Linum L. Species of the Section Linum (syn. Protolinum and Adenolinum), Russ. J. Genet., 2004, vol. 40, no. 2, pp. 193–196.
Samatadze, T.E., Muravenko, O.V., Klimakhin, G.I., and Zelenin, A.V., Intraspecific Chromosome Polymorphism in Matricaria chamomilla L. (syn. M. recutita L.) Studied by C-Banding Techniques, Russ. J. Genet., 1997, vol. 33, no. 1, pp. 111–113.
Samatadze, T.E., Muravenko, O.V., and Zelenin, A.V., Comparison of C-Banded Chromosomes in Karyotypes of Three Species of the Genus Matricaria L., Russ. J. Genet., 1998, vol. 34, no. 12, pp. 1469–1473.
Samatadze, T.E., Muravenko, O.V., Popov, K.V., and Zelenin, A.V., Genome Comparison of the Matricaria chamomilla L. Varieties by the Chromosome C- and ORBanding Patterns, Caryologia, 2001, vol. 54, no. 4, pp. 299–306.
Manoj, K., Friebe, B., Koul, A.K., and Gill, B.S., Origin of an Apparent B Chromosome by Mutation, Chromosome Fragmentation and Specific DNA Sequence Amplification, Chromosoma, 2002, vol. 111, pp. 332–340.
Cullis, C.A., Mechanisms and Control of Rapid Genomic Changes in Flax, Ann. Botany, 2005, vol. 95, pp. 201–206.
Miller, D.A., Dev, V.G., Tantravahi, R., and Miller, O.J., Suppression of Human Nucleolus Organizer Activity in Mouse-Human Somatic Hybrid Cell, Exp. Cell Res., 1976, vol. 101, pp. 235–243.
Miller, D.A., Tantravahi, R., Dev, V.G., et al., Frequency of Satellite Association of Human Chromosomes Is Correlated with Amount of Ag-Staining of the Nucleolus Organizer Region, Am. J. Hum. Genet., 1977, vol. 29, pp. 390–502.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © T.E. Samatadze, D.A. Zelenina, N.G. Shostak, A.A. Volkov, K.V. Popov, O.V. Rachinskaya, A.Yu. Borisov, I.A. Tihonovich, A.V. Zelenin, O.V. Muravenko, 2008, published in Genetika, 2008, Vol. 44, No. 12, pp. 1644–1651.
Rights and permissions
About this article
Cite this article
Samatadze, T.E., Zelenina, D.A., Shostak, N.G. et al. Comparative genome analysis in pea Pisum sativum L. Varieties and Lines with chromosomal and molecular markers. Russ J Genet 44, 1424–1430 (2008). https://doi.org/10.1134/S1022795408120065
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795408120065