Abstract
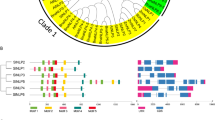
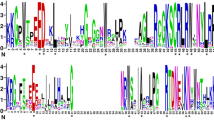
The NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) proteins play important roles in transporting substrates, such as nitrate, peptides, amino acids, dicarboxylates, malate, glucosinolates, indole acetic acid, abscisic acid, and jasmonic acid. However, there is limited information on the NPF genes in tomato (Solanum lycopersicum L.), in contrast to Arabidopsis. Our study aimed to reveal general information about tomato NPFs and to analyze the transcriptional responses of some members using plant nitrogen status. We identified 85 SlNPF genes, and a phylogenetic analysis organized them into nine major clades. Thirty motifs were found based on NPF amino acid sequence alignments. Chromosomal locations and gene duplication events of SlNPF family genes were also analyzed. An uneven distribution of SlNPF genes was discovered among tomato chromosomes. Twenty-five SlNPF genes resulted from whole-genome triplication (WGT)/segmental duplication in Solanaceae. Our results showed that ancient whole-genome triplication and tandem duplication mainly contributed to the expansion of the SlNPF genes. In the NPF family, 19 orthologous genes were identified between tomato and Arabidopsis, suggesting that at least 19 NPF genes were present in a common ancestor before Arabidopsis and tomato differentiated. In addition, we analyzed the expression patterns of the SlNPF family genes in various tomato tissues. We monitored 49 root-specific SlNPF genes that showed varied expression patterns under different N status. Among them, SINPF9, -34, ‑58 and -60 were significantly induced by both low and high levels of nitrate. Our findings provide a foundation for future research on this gene family.







Similar content being viewed by others
REFERENCES
O'Brien, J.A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., and Gutiérrez, R.A., Nitrate transport, sensing, and responses in plants, Mol. Plant, 2016, vol. 9, p. 837. https://doi.org/10.1016/j.molp.2016.05.004
Gojon, A., Krouk, G., Perrine-Walker, F., and Laugier, E., Nitrate transceptor(s) in plants, J. Exp. Bot., 2011, vol. 62, p. 2299. https://doi.org/10.1093/jxb/erq419
Krapp, A., David, L.C., Chardin, C., Girin, T., Marmagne, A., Leprince, A.S., Chaillou, S., Ferrario-Méry, S., Meyer, S., and Daniel-Vedele, F., Nitrate transport and signaling in Arabidopsis, J. Exp. Bot., 2014, vol. 65, p. 789. https://doi.org/10.1093/jxb/eru001
Wang, X., Cai, X., Xu, C., and Wang, Q., Identification and characterization of the NPF, NRT2, and NRT3 in spinach, Plant Physiol. Biochem., 2021, vol. 158, p. 297. https://doi.org/10.1016/j.plaphy.2020.11.017
Zhang, H., Li, S., Shi, M., Wang, S., Shi, L., Xu, F., and Ding, G., Genome-wide systematic characterization of the npf family genes and their transcriptional responses to multiple nutrient stresses in allotetraploid rapeseed, Int. J. Mol. Sci., 2020, vol. 21, p. 5947. https://doi.org/10.3390/ijms21175947
Wang, H., Wan, Y., Buchner, P., King, R., Ma, H., and Hawkesford, M.J., Phylogeny and gene expression of the complete NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in Triticum aestivum, J. Exp. Bot., 2020, vol. 71, p. 4531. https://doi.org/10.1093/jxb/eraa210
Drechsler, N., Courty, P.E., Brule, D., and Kunze, R., Identification of arbuscular mycorrhiza-inducible Nitrate Transporter 1/Peptide Transporter Family (NPF) genes in rice, Mycorrhiza, 2018, vol. 28, p. 93. https://doi.org/10.1007/s00572-017-0802-z
Chao, H., He, J., Cai, Q., Zhao, W., Fu, H., Hua, Y., Li, M., and Huang, J., The expression characteristics of NPF genes and their response to vernalization and nitrogen deficiency in rapeseed, Int. J. Mol. Sci., 2021, vol. 22, p. 4944. https://doi.org/10.3390/ijms22094944
Prabhala, B.K., Rahman, M., Nour-Eldin, H.H., Jorgensen, F.S., and Mirza, O., PTR2/POT/NPF transporters: what makes them tick? in Advances in Protein Chemistry and Structural Biology, Amsterdam: Elsevier, 2021, vol. 123, ch. 10, p. 219. https://doi.org/10.1016/bs.apcsb.2020.10.002.
Tsay, Y.F., Schroeder, J.I., Feldmann, K.A., and Crawford, N.M., The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter, Cell, 1993, vol. 72, p. 705. https://doi.org/10.1016/0092-8674(93)90399-b
Karim, S., Holmstrom, K.O., Mandal, A., Dahl, P., Hohmann, S., Brader, G., Palva, E.T., and Pirhonen, M., AtPTR3, a wound-induced peptide transporter needed for defence against virulent bacterial pathogens in Arabidopsis, Planta, 2007, vol. 225, p. 1431. https://doi.org/10.1007/s00425-006-0451-5
Nour-Eldin, H.H., Andersen, T.G., Burow, M., Madsen, S.R., Jorgensen, M.E., Olsen, C.E., Dreyer, I., Hedrich, R., Geiger, D., and Halkier, B.A., NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds, Nature, 2012, vol. 488, p. 531. https://doi.org/10.1038/nature11285
Mounier, E., Pervent, M., Ljung, K., Gojon, A., and Nacry, P., Auxin-mediated nitrate signaling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability, Plant Cell Environ., 2014, vol. 37, p. 162. https://doi.org/10.1111/pce.12143
Tal, I., Zhang, Y., Jorgensen, M.E., Pisanty, O., Barbosa, I.C., Zourelidou, M., Regnault, T., Crocoll C., Olsen, C.E., Weinstain, R., Schwechheimer, C., Halkier B.A., Nour-Eldin, H.H., Estelle, M., and Shani, E., The Arabidopsis NPF3 protein is a GA transporter, Nat. Commun., 2016, vol. 7, p. 11486. https://doi.org/10.1038/ncomms11486
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., Koshiba, T., Kamiya, Y., and Seo, M., Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor, Proc. Natl. Acad. Sci. U.S.A., 2012, vol. 109, p. 9653. https://doi.org/10.1073/pnas.1203567109
Liu, K.H. and Tsay, Y.F., Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation, EMBO J., 2003, vol. 22, p. 1005. https://doi.org/10.1093/emboj/cdg118
Hachiya, T. and Sakakibara, H., Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants, J. Exp. Bot., 2017, vol. 68, p. 2501. https://doi.org/10.1093/jxb/erw449
Dechorgnat, J., Nguyen, C.T., Armengaud, P., Jossier, M., Diatloff, E., Filleur, S., and Daniel-Vedele, F., From the soil to the seeds: the long journey of nitrate in plants, J. Exp. Bot., 2011, vol. 62, p. 1349. https://doi.org/10.1093/jxb/erq409
Wulff, N., Ernst, H.A., Jorgensen, M.E., Lambertz, S., Maierhofer, T., Belew, Z.M., Crocoll, C., Motawia, M.S., Geiger, D., Jørgensen, F.S., Mirza, O., and Nour-Eldin, H.H., An optimized screen reduces the number of GA transporters and provides insights into nitrate transporter 1/peptide transporter family substrate determinants, Front. Plant Sci., 2019, vol. 10, p. 1106. https://doi.org/10.3389/fpls.2019.01106
Weichert, A., Brinkmann, C., Komarova, N.Y., Dietrich, D., Thor, K., Meier, S., Grotemeyer, M.S., and Rentsch, D., AtPTR4 and AtPTR6 are differentially expressed, tonoplast-localized members of the peptide transporter/nitrate transporter 1 (PTR/NRT1) family, Planta, 2012, vol. 235, p. 311. https://doi.org/10.1007/s00425-011-1508-7
Tong, J., Walk, T.C., Han, P., Chen, L., Shen, X., Li, Y., Gu, C., Xie, L., Hu, X., Liao, X., and Qin, L., Genome-wide identification and analysis of high-affinity nitrate transporter 2 (NRT2) family genes in rapeseed (Brassica napus L.) and their responses to various stresses, BMC Plant Biol., 2020, vol. 20, p. 464. https://doi.org/10.1186/s12870-020-02648-1
Kiba, T., Feria-Bourrellier, A.B., Lafouge, F., Lezhneva, L., Boutet-Mercey, S., Orsel, M., Bréhaut, V., Miller, A., Daniel-Vedele, F., Sakakibara, H., and Krapp, A., The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants, Plant Cell, 2012, vol. 24, p. 245. https://doi.org/10.1105/tpc.111.092221
Liu, R., Jia, T., Cui, B., and Song, J., The expression patterns and putative function of nitrate transporter 2.5 in plants, Plant Signaling Behav., 2020, vol. 15, art. ID 1815980. https://doi.org/10.1080/15592324.2020.1815980
Finn, R.D., Coggill, P., Eberhardt, R.Y., Eddy, S.R., Mistry, J., Mitchell, A.L., Potter, S.C., Punta, M., Qureshi, M., Sangrador-Vegas, A., Salazar, G.A., Tate, J., and Bateman, A., The Pfam protein families database: towards a more sustainable future, Nucleic Acids Res., 2016, vol. 44, p. D279. https://doi.org/10.1093/nar/gkv1344
Eddy, S.R., Profile hidden Markov models, Bioinformatics, 1998, vol. 14, p. 755. https://doi.org/10.1093/bioinformatics/14.9.755
Katoh, K. and Standley, D.M., MAFFT multiple sequence alignment software version 7: improvements in performance and usability, Mol. Biol. Evol., 2013, vol. 30, p. 772. https://doi.org/10.1093/molbev/mst010
Keane, T.M., Creevey, C.J., Pentony, M.M., Naughton, T.J., and McLnerney, J.O., Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified, BMC Evol. Biol., 2006, vol. 6, p. 29. https://doi.org/10.1186/1471-2148-6-29
Bailey, T.L., Johnson, J., Grant, C.E., and Noble, W.S., The MEME suite, Nucleic Acids Res., 2015, vol. 43, p. W39. https://doi.org/10.1093/nar/gkv416
Wang, Y., Tang, H., Debarry, J.D., Tan, X., Li, J., Wang, X., Lee, T., Jin, H., Marler, B., Guo, H., Kissinger, J.C., and Paterson, A.H., MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity, Nucleic Acids Res., 2012, vol. 40, p. e49. https://doi.org/10.1093/nar/gkr1293
Suyama, M., Torrents, D., and Bork, P., PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments, Nucleic Acids Res., 2006, vol. 34, p. W609. https://doi.org/10.1093/nar/gkl315
Yang, Z., PAML 4: phylogenetic analysis by maximum likelihood, Mol. Biol. Evol., 2007, vol. 24, p. 1586. https://doi.org/10.1093/molbev/msm088
Chen, C., Chen, H., Zhang, Y., Thomas, H.R., Frank, M.H., He, Y., and Hia, R., TBtools: an integrative toolkit developed for interactive analyses of big biological data, Mol. Plant., 2020, vol. 13, p. 1194. https://doi.org/10.1016/j.molp.2020.06.009
Morales-Cruz, A., Amrine, K.C., Blanco-Ulate, B., Lawrence, D.P., Travadon, R., Rolshausen, P.E., Baumgartner, K., and Cantu, D., Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens, BMC Genomics, 2015, vol. 16, p. 469. https://doi.org/10.1186/s12864-015-1624-z
Zhao, T., Holmer, R., de Bruijn, S., Angenent, G.C., van den Burg, H.A., and Schranz, M.E., Phylogenomic synteny network analysis of MADS-box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation, Plant Cell, 2017, vol. 29, p. 1278. https://doi.org/10.1105/tpc.17.00312
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., Zheng, Y., and Wang, X., Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family, J. Exp. Bot., 2014, vol. 65, p. 1513. https://doi.org/10.1093/jxb/eru007
Jaillon, O., Aury, J.M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., Choisne, N., Aubourg, S., Vitulo, N., Jubin, C., Vezzi, A., Legeai, F., Hugueney, P., Dasilva, C., Horner, D., et al., The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla, Nature, 2007, vol. 449, p. 463. https://doi.org/10.1038/nature06148
Qiao, X., Yin, H., Li, L., Wang, R., Wu, J., Wu, J., and Zhang, S., Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri), Front. Plant Sci., 2018, vol. 9, p. 161. https://doi.org/10.3389/fpls.2018.00161
Renny-Byfield, S., Gallagher, J.P., Grover, C.E., Sz-adkowski, E., Page, J.T, Udall, J.A., Wang. X., Paterson, A.H., and Wendel, J.F., Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence, Genome Biol. Evol., 2014, vol. 6, p. 559. https://doi.org/10.1093/gbe/evu037
Liu, Z., Coulter, J.A., Li, Y., Zhang, X., Meng, J., Zhang, J., and Liu, Y., Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.), Int. J. Biol. Macromol., 2020, vol. 153, p. 327. https://doi.org/10.1016/j.ijbiomac.2020.03.022
Taylor, S., Pieri, K., Nanni, P., Tica, J., Barratt, J., and Didangelos, A., Phosphatidylethanolamine binding protein-4 (PEBP4) is increased in IgA nephropathy and is associated with IgA-positive B-cells in affected kidneys, J. Autoimmun., 2019, vol. 105, art. ID 102309. https://doi.org/10.1016/j.jaut.2019.102309
ACKNOWLEDGMENTS
We thank members of the ZY laboratory for discussions and comments on the manuscript. We thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This work was financially supported by the Scientific and Technological Project of Jinhua City (project nos. 2021-2-027 and 2021-2-019).
Author information
Authors and Affiliations
Contributions
X. Liu and Y. Gao contributed equally to this work.
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflicts of interest.
Statement on the welfare of humans or animals. This article does not contain any studies involving humans or animals performed by any of the authors.
Additional information
Abbreviations: NPF—NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER; WGT—whole-genome triplication; NRTs—nitrate transporters; PTR—peptide transporter; AP2/ERF—APETALA2/ethylene-responsive factor; WGD—whole-genome duplication.
Supplementary Information
Rights and permissions
About this article
Cite this article
Liu, X., Gao, Y., Li, K. et al. Complex Phylogeny and Expression Patterns of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER Family Genes in Tomato. Russ J Plant Physiol 69, 47 (2022). https://doi.org/10.1134/S1021443722030086
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1134/S1021443722030086