Abstract
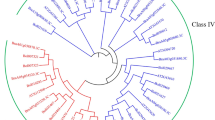
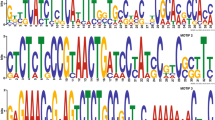
Plant chitinases play a key role in conferring resistance to environmental stresses, including attack by fungal pathogens. In the present study, we employed rapid amplification of cDNA ends (RACE) to identify five chitinase genes in Populus canadensis Moench. Sequence alignment revealed that these genes belong to five subfamilies of chitinase genes. The full-length cDNAs of these genes ranged in size from 991 to 1358 bp and encoded proteins with mol wts from 29.5 to 40.3 kD. Five genes were grouped into three major clades based on amino acid sequences of encoded proteins. Exon-intron gene structure and protein domain analysis further supported the designation. A three-dimensional structure comparison showed the high similarity between five P. canadensis chitinases and three well-studied chitinases from other species. The expression levels of all five genes were up-regulated during Populus infection with the pathogenic fungus Marssonina brunnea, and four of them were highly induced by salt and drought stresses. Furthermore, such factors as elicitors, wounding, and low temperature also elevated the expression of these chitinase genes to varying extents. We postulated that these chitinase genes may be involved in pathways of the defense against fungal infection and function in response to various abiotic stresses.
Similar content being viewed by others
Abbreviations
- CTAB:
-
cetyltrimethylammonium bromide
- RACE:
-
rapid amplification of cDNA ends
- RT-PCR:
-
reverse transcription-polymerase chain reaction
References
Cohen-Kupiec, R. and Chet, I., The Molecular Biology of Chitin Digestion, Curr. Opin. Biotechnol., 1998, vol. 9, pp. 270–277.
Kasprzewska, A., Plant Chitinases — Regulation and Function, Cell Mol. Biol. Lett., 2003, vol. 8, pp. 809–824.
Sharma, N., Sharma, K., Gaur, R., and Gupta, V., Role of Chitinase in Plant Defense, Asian. J. Biochem., 2011, vol. 6, pp. 29–37.
Ubhayasekera, W., Structure and Function of Chitinases from Glycoside Hydrolase Family 19, Polymer Int., 2011, vol. 60, pp. 890–896.
Shimono, K., Matsuda, H., and Kawamukai, M., Functional Expression of Chitinase and Chitosanase, and Their Effects on Morphologies in the Yeast Schizosaccharomyces pombe, BioSci. Biotechnol. Biochem., 2002, vol. 66, pp. 1143–1147.
Zhang, J., Zhang, X., Arakane, Y., Muthukrishnan, S., Kramer, K.J., Ma, E., and Zhu, K.Y., Comparative Genomic Analysis of Chitinase and Chitinase-Like Genes in the African Malaria Mosquito (Anopheles gambiae), PLoS One, 2011, vol. 6, p. e19899.
Santos, P., Fortunato, A., Ribeiro, A., and Pawlowski, K., Chitinases in Root Nodules, Plant Biotechnol., 2008, vol. 25, pp. 299–307.
Passarinho, P.A. and de Vries, S.C., Arabidopsis Chitinases: A Genomic Survey, The Arabidopsis Book, 2002, pp. 1–25.
Kim, S.H., Choi, H.S., Cho, Y.C., and Kim, S.R., Cold-Responsive Regulation of a Flower-Preferential Class III Peroxidase Gene, ospox1, in Rice (Oryza sativa L.), J. Plant Biol., 2012, vol. 55, pp. 123–131.
Brameld, K.A. and Goddard, W.A., The Role of Enzyme Distortion in the Single Displacement Mechanism of Family 19 Chitinases, Proc. Natl. Acad. Sci. USA, 1998, vol. 95, pp. 4276–4281.
Van Aalten, D., Komander, D., Synstad, B., Gåseidnes, S., Peter, M., and Eijsink, V., Structural Insights into the Catalytic Mechanism of a Family 18 Exo-Chitin, Proc. Natl. Acad. Sci. USA,2001, vol. 98, pp. 8979–8984.
Hamel, F., Boivin, R., Tremblay, C., and Bellemare, G., Structural and Evolutionary Relationships among Chitinases of Flowering Plants, J. Mol. Evol., 1997, vol. 44, pp. 614–624.
Graham, L.S. and Sticklen, M.B., Plant Chitinases, Can. J. Bot., 1994, vol. 72, pp. 1057–1083.
Nandakumar, R., Babu, S., Kalpana, K., Raguchander, T., Balasubramanian, P., and Samiyappan, R., Agrobacterium-Mediated Transformation of Indica Rice with Chitinase Gene for Enhanced Sheath Blight Resistance, Biol. Plant., 2007, vol. 51, pp. 142–148.
Salami, S., Ebadi, A., Naghavi, M., and Dry, I., Cloning and Functional Characterization of a Class III Chitinase Gene from Grapevine: Inhibition of Fungal Growth by Recombinant VvChiF III, Afr. J. Biotechnol., 2008, vol. 7, pp. 4400–4405.
Naher, L., Ho, C.L., Tan, S.G., Yusuf, U.K., and Abdullah, F., Cloning of Transcripts Encoding Chitinases from Elaeis guineensis Jacq. and Their Expression Profiles in Response to Fungal Infections, Physiol. Mol. Plant Pathol., 2011, vol. 76, pp. 96–103.
Tuskan, G.A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., Putnam, N., Ralph, S., Rombauts, S., Salamov, A., Schein, J., Sterck, L., Aerts, A., Bhalerao, R.R., Bhalerao, R.P., Blaudez, D., Boerjan, W., Brun, A., Brunner, A., Busov, V., Campbell, M., Carlson, J., Chalot, M., Chapman, J., Chen, G.L., Cooper, D., Coutinho, P.M., Couturier, J., Covert, S., Cronk, Q., Cunningham, R., Davis, J., Degroeve, S., Dejardin, A., Depamphilis, C., Detter, J., Dirks, B., Dubchak, I., Duplessis, S., Ehlting, J., Ellis, B., Gendler, K., Goodstein, D., Gribskov, M., Grimwood, J., Groover, A., Gunter, L., Hamberger, B., Heinze, B., Helariutta, Y., Henrissat, B., Holligan, D., Holt, R., Huang, W., Islam-Faridi, N., Jones, S., Jones-Rhoades, M., Jorgensen, R., Joshi, C., Kangasjärvi, J., Karlsson, J., Kelleher, C., Kirkpatrick, R., Kirst, M., Kohler, A., Kalluri, U., Larimer, F., Leebens-Mack, J., Leplé, J.C., Locascio, P., Lou, Y., Lucas, S., Martin, F., Montanini, B., Napoli, C., Nelson, D.R., Nelson, C., Nieminen, K., Nilsson, O., Pereda, V., Peter, G., Philippe, R., Pilate, G., Poliakov, A., Razumovskaya, J., Richardson, P., Rinaldi, C., Ritland, K., Rouzé, P., Ryaboy, D., Schmutz, J., Schrader, J., Segerman, B., Shin, H., Siddiqui, A., Sterky, F., Terry, A., Tsai, C.J., Uberbacher, E., Unneberg, P., Vahala, J., Wall, K., Wessler, S., Yang, G., Yin, T., Douglas, C., Marra, M., Sandberg, G., van de Peer, Y., and Rokhsar, D., The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray), Science, 2006, vol. 313, pp. 1596–1604.
Chung, H.S., Koo, A.J., Gao, X., Jayanty, S., Thines, B., Jones, A.D., and Howe, G.A., Regulation and Function of Arabidopsis JASMONATE ZIM-Domain Genes in Response to Wounding and Herbivory, Plant Physiol., 2008, vol. 146, pp. 952–964.
Cheng, Q., Cao, Y., Jiang, C., Xu, L., Wang, M., Zhang, S., and Huang, M., Identifying Secreted Proteins of Marssonina brunnea by Degenerate PCR, Proteomics, 2010, vol. 10, pp. 2406–2417.
Xu, M., Zhang, B., Yao, H.S., and Huang, M.R., Isolation of High Quality RNA and Molecular Manipulations with Various Tissues of Populus, Russ. J. Plant Physiol., 2009, vol. 56, pp. 716–719.
Zhang, Y., I-Tasser Server for Protein 3D Structure Prediction, BMC Bioinformatics, 2008, vol. 9, p. 40.
Roy, A., Kucukural, A., and Zhang, Y., I-Tasser: A Unified Platform for Automated Protein Structure and Function Prediction, Nat. Protoc., 2010, vol. 5, pp. 725–738.
Holm, L. and Park, J., Dalilite Workbench for Protein Structure Comparison, Bioinformatics, 2000, vol. 16, pp. 566–567.
Kezuka, Y., Kojima, M., Mizuno, R., Suzuki, K., Watanabe, T., and Nonaka, T., Structure of Full-Length Class I Chitinase from Rice Revealed by X-Ray Crystallography and Small-Angle X-Ray Scattering, Proteins, 2010, vol. 78, pp. 2295–2309.
Ohnuma, T., Numata, T., Osawa, T., Mizuhara, M., Lampela, O., Juffer, A.H., Skriver, K., and Fukamizo, T., A Class V Chitinase from Arabidopsis thaliana: Gene Responses, Enzymatic Properties, and Crystallographic Analysis, Planta, 2011, vol. 234, pp. 123–137.
Hennig, M., Jansonius, J.N., Terwisscha van Scheltinga, A.C., Dijkstra B.W., and Schlesier, B., Crystal Structure of Concanavalin B at 1.65 A Resolution. An “Inactivated” Chitinase from Seeds of Canavalia ensiformis, J. Mol. Biol., 1995, vol. 254, pp. 237–246.
Author information
Authors and Affiliations
Corresponding authors
Additional information
This text was submitted by the authors in English.
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Jiang, C., Song, J., Huang, R. et al. Cloning and expression analysis of Chitinase genes from Populus canadensis . Russ J Plant Physiol 60, 396–403 (2013). https://doi.org/10.1134/S1021443713030072
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443713030072