Abstract
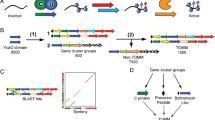
Nonribosomal peptides play an important role in the vital activity of bacteria and have an extremely broad field of biological activity. In particular, they act as antibiotics, toxins, surfactants, siderophores, and also perform a number of other specific functions. Biosynthesis of these molecules does not occur on ribosomes but by special enzymes that form gene clusters in bacterial genomes. We hypothesized that the presence of nonribosomal peptide synthesis pathways is a specific feature of bacterial metabolism, which may affect other vital processes of the cell, including translational ones. This work was the first to show the relationship between the translation regulation mechanism of protein-coding genes in bacteria, which is largely determined by the efficiency of translation elongation, and the presence of gene clusters in the genomes for the biosynthesis of nonribosomal peptides. Bioinformatic analysis of the translation elongation efficiency of protein-coding genes was performed in 11 679 bacterial genomes, some of which contained gene clusters of nonribosomal peptide biosynthesis and some of which did not. The analysis showed that bacteria whose genomes contained clusters of nonribosomal peptide biosynthetic genes and those without such gene clusters differ significantly in the molecular mechanisms that ensure translation efficiency. Thus, among microorganisms whose genomes contain gene clusters of nonribosomal peptide synthetases, a significantly smaller part of them is characterized by optimized regulation of the number of local inverted repeats, while most of them have genomes optimized by the averaged energy of inverted repeats studs in mRNA and additionally by codon composition. Our results suggest that the presence of nonribosomal peptide biosynthetic pathways in bacteria may influence the structure of the overall bacterial metabolism, which is also expressed in the specific mechanisms of ribosomal protein biosynthesis.






Similar content being viewed by others
REFERENCES
Caboche S., Pupin M., Leclère V., Fontaine A., Jacques P., Kucherov G. 2008. NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 36, 326–331. https://doi.org/10.1093/nar/gkm792
Süssmuth R.D., Mainz A. 2017. Nonribosomal peptide synthesis—principles and prospects. Angew. Chemie, Int. Ed. 56, 3770–3821.
Kim H.U., Blin K., Lee S.Y., Weber T. 2017. Recent development of computational resources for new antibiotics discovery. Curr. Opin. Microbiol. 39, 113–120.
Blin K., Shaw S., Kautsar S.A., Medema M.H., Weber T. 2021. The antiSMASH database version 3: increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 49 (D1), D639‒D643.
Likhoshvai V.A., Matushkin Yu.G. 2000. Nucleotide composition-based prediction of gene expression efficacy. Mol. Biol. (Moscow). 34, 345–350.
Likhoshvai V.A., Matushkin Yu.G. 2002. Differentiation of single-cell organisms according to elongation stages crucial for gene expression efficacy. FEBS Lett. 516. 87–92.
Sokolov V. S., Zuraev B. S., Lashin S. A., Matushkin Yu. G. 2014EloE—a web application for evaluating the efficiency of gene translation elongation. Vavilov. Zh. Genet. Sel. 18, 904–909.
Korenskaia A.E., Matushkin Y.G., Lashin S.A., Klimenko A.I. 2022. Bioinformatic assessment of factors affecting the correlation between protein abundance and elongation efficiency in Prokaryotes. Int. J. Mol. Sci. 23 (19), 11996. https://doi.org/10.3390/ijms231911996
Blin K., Medema M.H., Kottmann R., Lee S.Y., Weber T. 2017. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 45, D555–D559. https://doi.org/10.1093/nar/gkw9601
Filzmoser P., Hron K., Templ M. 2018. Applied compositional data analysis with worked examples. In: Statistics. Springer Ser., Nature Switzerland AG, Cham, Switzerland. ISBN 978-3-319-96420-1.
Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Chen I.M.A., Kyrpides N.C., Reddy T.B.K. 2019. Genomes OnLine database (GOLD) v.7: Updates and new features. Nucleic Acids Res. 47 (D1), D649–D659. https://doi.org/10.1093/nar/gky977
Turner D.H., Sugimoto N. 1988. RNA structure prediction. Annu. Rev. Biophys. Biophys. Chem. 17, 167–192.
Sharp P.M., Li W.H. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295.
Filzmoser P., Hron K., Reimann C. 2007. Principal component analysis for compositional data with outliers. Environmetrics. 20, 621–632.
Funding
This work was supported by the Russian Foundation for Basic Research (project nos. 17-00-00470 (K) and 17-00-00462). Data were processed using the computing resources of the Bioinformatika (Bioinformatics?) Core Facility with the support of the budgetary project no. FWNR-2022-0020.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by M. Batrukova
Abbreviations: NRPs, nonribosomal peptides; NRPS, nonribosomal peptide synthetase; EEI, elongation efficiency index.
Rights and permissions
About this article
Cite this article
Klimenko, A.I., Lashin, S.A., Kolchanov, N.A. et al. Molecular Mechanisms to Optimize Gene Translation Elongation Differ Significantly in Bacteria with and without Nonribosomal Peptides. Mol Biol 57, 155–164 (2023). https://doi.org/10.1134/S0026893323020115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893323020115