Abstract
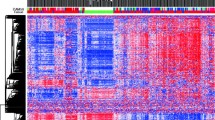
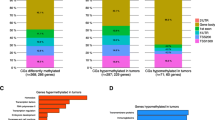
For the first time, the epigenetic status of breast benign proliferative processes, malignant breast tumors, and metastases to regional lymph nodes has been studied using the GoldenGate Cancer Panel I DNA methylation microarray (Illumina, United States). The functional groups of differentially methylated genes were identified in each set of samples. The aberrant methylation of genes that regulate cell proliferation and mobility was found in the samples of benign proliferative breast processes. The aberrant methylation of genes responsible for cell differentiation and proliferation, as well as protein phosphorylation and cell mobility, was observed in the samples of malignant breast tumors. The differential methylation of the genes that regulate cell adhesion, the formation of anatomical structures, angiogenesis, immune response, signal transduction, and protein phosphorylation were found in samples with metastases to regional lymph nodes compared to the unaltered breast epithelium. It was found that tissues that range from benign proliferative processes and metastases to regional lymph nodes were generally characterized by a relatively lower level of epigenetic variability compared to the tissues of the primary tumor.
Similar content being viewed by others
Abbreviations
- BC:
-
breast cancer
- BPP:
-
benign proliferative processes
- RLN:
-
regional lymph nodes
References
Zemlyakova V.V., Zhevlova A.I., Strelnikov V.V., Lyubchenko L.N., Shabanov M.A., Vishnevskaya Ya.V., Tretyakova V.A., Zaletayev D.V., Nemtsova M.V. 2003. Abnormal methylation of several tumor suppressor genes in sporadic breast cancer. Mol. Biol. (Moscow). 37, 591–597.
Zaletaev D.V., Nemtsova M.V., Strelnikov V.V., Babenko O.V., Vasil’ev E.V., Zemlyakova V.V., Zhevlova A.I., Drozd O.V. 2004. Diagnostics of epigenetic alterations in hereditary and oncological disorders. Mol. Biol. (Moscow). 38, 174–182.
Esteller M. 2008. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159.
Feinberg A.P. 2010. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 456, 13–21.
O’Riain C., O’Shea D.M., Yang Y., et al. 2009. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 23, 1858–1866.
Costello J.F., Plass C. 2001. Methylation matters. J. Med. Genet. 38, 285–303.
Ehrlich M. 2002. DNA methylation in cancer: Too much, but also too little. Oncogene. 21, 5400–5413.
Plass C., Soloway P.D. 2002. DNA methylation, imprinting, and cancer. Eur. J. Hum. Genet. 10, 6–16.
Chien W., O’Kelly J., Lu D., et al. 2011. Expression of connective tissue growth factor (CTGF/CCN2) in breast cancer cells is associated with increased migration and angiogenesis. Int. J. Oncol. 38, 1741–1747.
Getsios S., Amargo E.V., Dusek R.L., et al. 2004. Coordinated expression of desmoglein 1 and desmocollin 1 regulates intercellular adhesion. Differentiation. 72, 419–433.
Hartman A.R. 2005. The problems with risk selection: Scientific and psychosocial aspects. Recent Results Cancer Res. 166, 125–144.
Nagaraju G.P., Sharma D. 2011. Anti-cancer role of SPARC, an inhibitor of adipogenesis. Cancer Treat. Rev. 37, 559–566.
Chen D., Yoo B.K., Santhekadur P.K., et al. 2011. Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin. Cancer Res. 17, 6693–6701.
Zhen L., Lei Y., Dong-Xu C., et al. 2007. Methylation status and protein expression of adenomatous polyposis coli (APC) gene in breast cancer. Chinese J. Cancer. 26, 586–590.
Said N., Sanchez-Carbayo M., Smith S.C., Theodorescu D. 2012. RhoGDI2 suppresses lung metastasis in mice by reducing tumor versican expression and macrophage infiltration. J. Clin. Invest. 122, 1503–1518.
Kim S., Takahashi H., Lin W., et al. 2009. Carcinomaproduced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 457, 102–107.
Menashe I., Figueroa J.D., Garcia-Closas M., et al. 2012. Large-scale pathway-based analysis of bladder cancer genome-wide association data from five studies of European background. PLOS ONE. 7, e29396.
Skryabin N.A., Lebedev I.N., Tolmacheva E.N., Vtorushin S.V., Slonimskaya E.M., Cherdyntseva N.V. 2011. Epigenetic aspects of early lymphogenic metastasis in breast cancer. Vopr. Onkol. 57, 717–721.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © N.A. Skryabin, E.N. Tolmacheva, I.N. Lebedev, M.V. Zavyalova, E.M. Slonimskaya, N.V. Cherdyntseva, 2013, published in Molekulyarnaya Biologiya, 2013, Vol. 47, No. 2, pp. 302–310.
Rights and permissions
About this article
Cite this article
Skryabin, N.A., Tolmacheva, E.N., Lebedev, I.N. et al. Dynamics of aberrant methylation of functional groups of genes in progression of breast cancer. Mol Biol 47, 267–274 (2013). https://doi.org/10.1134/S0026893313020131
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893313020131