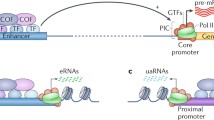
Abstract
Complete DNA sequences of many animal genomes combined with the information on new classes of regulatory elements within the regions conventionally viewed as intergenic led to a revision of the concept of the genome as a linear arrangement of genes with their promoters and distinct intergenic regions. Instead, there emerged a concept of the so-called transcriptional landscape with almost no boundaries between what was commonly considered to be genes. The concept of the core promoter as the main cis-acting transcriptional element also changed dramatically. Knowledge of the mechanisms sustaining the function of this central element of the cell transcription system underlies the understanding of metazoan transcription in general. The review attempts to summarize the data obtained for core promoters in the past decade and to trace the evolution of the core promoter concept. This evolution led finally to the understanding that core promoters play an active role in transcription regulation rather than just provide a passive scaffold for assembling preinitiation transcription complexes.
Similar content being viewed by others
References
Domansky A.N., Kopantzev E.P., Snezhkov E.V., Lebedev Y.B., Leib-Mosch C., Sverdlov E.D. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472, 191–195.
Sugimoto M., Miyata S. 2002. Functional property of von Willebrand factor under flowing blood. Int. J. Hematol. 75, 19–24.
Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P. Myers R.M. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14, 62–66.
Piontkivska H., Yang M.Q., Larkin D.M., Lewin H.A., Reecy J., Elnitski L. 2009. Cross-species mapping of bidirectional promoters enables prediction of unannotated 5′ UTRs and identification of species-specific transcripts. BMC Genomics. 10, 189.
Berretta J., Morillon A. 2009. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 10, 973–982.
Kapranov P., Willingham A.T., Gingeras T.R. 2007. Genome-wide transcription and the implications for genomic organization. Nature Rev. Genet. 8, 413–423.
Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D.A. 2007. Mammalian RNA polymerase II core promoters: Insights from genomewide studies. Nature Rev. Genet. 8, 424–436.
Jacquier A. 2009. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nature Rev. Genet. 10, 833–844.
Butler J.E., Kadonaga J. T. 2002. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes Dev. 16, 2583–2592.
Casamassimi A., Napoli C. 2007. Mediator complexes and eukaryotic transcription regulation: An overview. Biochimie. 89, 1439–1446.
Hahn S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nature Struct. Mol. Biol. 11, 394–403.
Kornberg R.D. 2007. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA. 104, 12955–12961.
Muller F., Demeny M.A., Tora L. 2007. New problems in RNA polymerase II transcription initiation: Matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 282, 14685–14689.
Thomas M.C., Chiang C.M. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178.
Venters B.J., Pugh B.F. 2009. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 44, 117–141.
Maston G.A., Evans S.K., Green M.R. 2006. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7, 29–59.
Sikorski T.W., Buratowski S. 2009. The basal initiation machinery: Beyond the general transcription factors. Curr. Opin. Cell Biol. 21, 344–351.
Core L.J., Waterfall J.J., Lis J. T. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 322, 1845–1848.
Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nature Genet. 39, 1235–1244.
Arndt K.M., Kane C.M. 2003. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 19, 543–550.
Hargreaves D.C., Horng T., Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 138, 129–145.
West S., Proudfoot N.J. 2009. Transcriptional termination enhances protein expression in human cells. Mol. Cell. 33, 354–364.
Fuda N.J., Ardehali M.B., Lis J. T. 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 461, 186–192.
Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A. Sharp P.A. 2008. Divergent transcription from active promoters. Science. 322, 1849–1851.
Taft R.J., Glazov E.A., Cloonan N., Simons C., Stephen S., Faulkner G.J., Lassmann T., Forrest A.R., Grimmond S.M., Schroder K., Irvine K., Arakawa T., Nakamura M., Kubosaki A., Hayashida K., Kawazu C., Murata M., Nishiyori H., Fukuda S., Kawai J., Daub C.O., Hume D.A., Suzuki H., Orlando V., Carninci P., Hayashizaki Y., Mattick J. S. 2009. Tiny RNAs associated with transcription start sites in animals. Nature Genet. 41, 572–578.
He Y., Vogelstein B., Velculescu V.E., Papadopoulos N., Kinzler K.W. 2008. The antisense transcriptomes of human cells. Science. 322, 1855–1857.
Buratowski S. 2008. Transcription. Gene expression-where to start? Science. 322, 1804–1805.
Ross J., Bottardi S., Bourgoin V., Wollenschlaeger A., Drobetsky E., Trudel M., Milot E. 2009. Differential requirement of a distal regulatory region for pre-initiation complex formation at globin gene promoters. Nucleic Acids Res. 37, 5295–5308.
Szutorisz H., Dillon N., Tora L. 2005. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem. Sci. 30, 593–599.
Juven-Gershon T., Hsu J.Y., Theisen J.W. Kadonaga J.T. 2008. The RNA polymerase II core promoter: The gateway to transcription. Curr. Opin. Cell Biol. 20, 253–259.
Kawaji H., Kasukawa T., Fukuda S., Katayama S., Kai C., Kawai J., Carninci P. Hayashizaki Y. 2006. CAGE Basic/Analysis Databases: The CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 34, D632–D636.
de Hoon M., Hayashizaki Y. 2008. Deep cap analysis gene expression (CAGE): Genome-wide identification of promoters, quantification of their expression, and network inference. Biotechniques. 44, 627–628, 630, 632.
Gershenzon N.I., Ioshikhes I. P. 2005. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 21, 1295–1300.
Suzuki M.M., Bird A. 2008. DNA methylation landscapes: Provocative insights from epigenomics. Nature Rev. Genet. 9, 465–476.
Davuluri R.V., Grosse I., Zhang M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nature Genet. 29, 412–417.
Saxonov S., Berg P., Brutlag D.L. 2006. A genomewide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA. 103, 1412–1417.
Ramirez-Carrozzi V.R., Braas D., Bhatt D.M., Cheng C.S., Hong C., Doty K.R., Black J.C., Hoffmann A., Carey M., Smale S. T. 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 138, 114–128.
Cairns B.R. 2009. The logic of chromatin architecture and remodelling at promoters. Nature. 461, 193–198.
Jiang C., Pugh B.F. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nature Rev. Genet. 10, 161–172.
Singh H. 2009. Teeing up transcription on CpG islands. Cell. 138, 14–16.
Smale S.T., Kadonaga J. T. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72, 449–479.
Baek D., Davis C., Ewing B., Gordon D., Green P. 2007. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 17, 145–155.
Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W., Ching K.A., Antosiewicz-Bourget J.E., Liu H., Zhang X., Green R.D., Lobanenkov V.V., Stewart R., Thomson J.A., Crawford G.E., Kellis M., Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 459, 108–112.
Davuluri R.V., Suzuki Y., Sugano S., Plass C., Huang T.H. 2008. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 24, 167–177.
Singer G.A., Wu J., Yan P., Plass C., Huang T.H., Davuluri R. V. 2008. Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genomics. 9, 349.
Weigmann A., Corbeil D., Hellwig A. Huttner W.B. 1997. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of nonepithelial cells. Proc. Natl. Acad. Sci. USA. 94, 12425–12430.
Shmelkov S.V., Jun L., St Clair R., McGarrigle D., Derderian C.A., Usenko J.K., Costa C., Zhang F., Guo X. Rafii S. 2004. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein AC133. Blood. 103, 2055–2061.
Pleshkan V.V., Vinogradova T.V., Sverdlov E.D. 2008. Methylation of the prominin 1 TATA-less main promoters and tissue specificity of their transcript content. Biochim. Biophys. Acta. 1779, 599–605.
Carninci P., Kasukawa T., Katayama S., et al. 2005. The transcriptional landscape of the mammalian genome. Science. 309, 1559–1563.
Narlikar G.J., Fan H.Y., Kingston R.E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 108, 475–487.
Becker P.B., Horz W. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247–273.
Saha A., Wittmeyer J., Cairns B.R. 2006. Chromatin remodelling: The industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7, 437–447.
Stein A., Takasuka T.E., Collings C.K. 2010. Are nucleosome positions in vivo primarily determined by histone-DNA sequence preferences? Nucleic Acids Res. 38, 709–719.
Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20, 90–100.
Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 36, 900–905.
Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 309, 626–630.
Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., Tomsho L.P., Qi J., Glaser R.L., Schuster S.C., Gilmour D.S., Albert I., Pugh B.F. 2008. Nucleosome organization in the Drosophila genome. Nature. 453, 358–362.
Tirosh I., Barkai N. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 18, 1084–1091.
Field Y., Kaplan N., Fondufe-Mittendorf Y., Moore I.K., Sharon E., Lubling Y., Widom J., Segal E. 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 4, e1000216.
Basehoar A.D., Zanton S.J., Pugh B.F. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 116, 699–709.
Huisinga K.L., Pugh B.F. 2004. A genome-wide house-keeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 13, 573–585.
Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science. 315, 1405–1408.
Rufiange A., Jacques P.E., Bhat W., Robert F., Nourani A. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 27, 393–405.
Mito Y., Henikoff J.G., Henikoff S. 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science. 315, 1408–1411.
Lee S., Kohane I., Kasif S. 2005. Genes involved in complex adaptive processes tend to have highly conserved upstream regions in mammalian genomes. BMC Genomics. 6, 168.
Tirosh I., Weinberger A., Carmi M., Barkai N. 2006. A genetic signature of interspecies variations in gene expression. Nature Genet. 38, 830–834.
Liang H., Lin Y.S., Li W.H. 2008. Fast evolution of core promoters in primate genomes. Mol. Biol. Evol. 25, 1239–1244.
Keightley P.D., Lercher M.J., Eyre-Walker A. 2005. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 3, e42.
Taylor M.S., Kai C., Kawai J., Carninci P., Hayashizaki Y., Semple C.A. 2006. Heterotachy in mammalian promoter evolution. PLoS Genet. 2, e30.
Okamura K., Nakai K. 2008. Retrotransposition as a source of new promoters. Mol. Biol. Evol. 25, 1231–1238.
Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.L., Ruan Y., Wei C.L., Ng H.H., Liu E.T. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762.
Kazazian H.H., Jr. 2004. Mobile elements: Drivers of genome evolution. Science. 303, 1626–1632.
Cordaux R., Batzer M.A. 2009. The impact of retrotransposons on human genome evolution. Nature Rev. Genet. 10, 691–703.
Sverdlov E.D. 2000. Retroviruses and primate evolution. Bioessays. 22, 161–171.
Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nature Rev. Genet. 9, 397–405.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © E.D. Sverdlov, T.V. Vinogradova, 2010, published in Molekulyarnaya Biologiya, 2010, Vol. 44, No. 5, pp. 773–785.
Rights and permissions
About this article
Cite this article
Sverdlov, E.D., Vinogradova, T.V. Core promoters as an example of the effect of whole-genome information on the evolution of views on molecular mechanisms of vital activity. Mol Biol 44, 682–692 (2010). https://doi.org/10.1134/S002689331005002X
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002689331005002X