Abstract
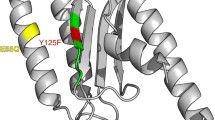
Differential scanning calorimetry (DSC) was used to study thermal denaturation of the human class 1 translation termination factor eRF1 and its mutants. Free energy changes caused by amino acid substitutions in the N domain were computed for eRF1. The melting of eRF1, consisting of three domains, proved to be cooperative. The thermostability of eRF1 was not affected by certain substitutions and was slightly increased by certain others. The corresponding residues were assumed to play no role in maintaining the eRF1 structure, which agreed with the published X-ray data. In these mutants (E55D, Y125F, N61S, E55R, E55A, N61S + S64D, C127A, and S64D), a selective loss of the capability to induce hydrolysis of peptidyl-tRNA in the ribosomal P site in the presence of a stop codon was not associated with destabilization of their spatial structure. Rather, the loss was due to local changes in the stereochemistry of the side groups of the corresponding residues in functionally important sites of the N domain. Two amino acid residues of the N domain, N129 and F131, proved to play an important role in the structural stability of eRF1 and to affect the selective recognition of mRNA stop codons in the ribosome. The recognition of the UAG and UAA stop codons in vitro was more tightly associated with the stability of the spatial structure of eRF1 as compared with that of the UGA stop codon.
Similar content being viewed by others
References
Kisselev L.L., Ehrenberg M., Frolova L.Yu. 2003. Termination of translation: interplay of mRNA, rRNAs, and release factors. EMBO J. 22, 175–182.
Kisselev L.L. 2003. Class-1 translation termination factors are functional analogs of aminoacyl-tRNAs. Mol. Biol. 37, 931–943.
Nakamura Y., Ito K. 2003. Making sense of mimic in translation termination. Trends Biochem. Sci. 28, 99–105.
Inge-Vechtomov S, Zhouravleva G, Philippe M. 2003. Eukaryotic release factors (eRFs) history. Biol. Cell. 95(3–4), 195–209.
Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. 2000. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 100, 311–321.
Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. 2000. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 6, 1236–1247.
Frolova L., Seit-Nebi A., Kisselev L. 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 8, 129–136.
Ito K., Frolova L., Seit-Nebi A., Karamyshev A., Kisselev L., Nakamura Y. 2002. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl. Acad. Sci. USA. 99, 8494–8499.
Seit-Nebi A., Frolova L., Kisselev L. 2002. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 3, 881–886.
Frolova L.Y., Tsivkovskii R.Y., Sivolobova G.F., Oparina N.Y., Serpinsky O.I., Blinov V.M., Tatkov S.I., Kisselev L.L. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 5, 1014–1020.
Seit-Nebi A., Frolova L., Justesen J., Kisselev L. 2001. Class-1 translation termination factors: Invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 29, 3982–3987.
Ito K., Ebihara K., Nakamura Y. 1998. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 4, 958–972.
Merkulova T.I., Frolova L.Y., Lazar M., Camonis J., Kisselev L.L. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443, 41–47.
Ebihara K, Nakamura Y. 1999. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA. 5, 739–750.
Chavatte L., Frolova L., Kisselev L., Favre A. 2001. The polypeptide chain release factor eRF1 specifically contacts the s(4)UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 268, 2896–2904.
Bulygin K.N., Repkova M.N., Ven’yaminova A.G., Graifer D.M., Karpova G.G., Frolova L.Yu., Kisselev L.L. 2002. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett. 514, 96–101.
Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO. 21, 5302–5311.
Chavatte L., Kervestin S., Favre A., Jean-Jean O. 2003. Stop codon selection in eukaryotic translation termination: Comparison of the discriminating potential between human and ciliate eRF1s. EMBO. 22, 1644–1653.
Bulygin K., Chavatte L., Frolova L., Karpova G., Favre A. 2005. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30, and S15. Biochemistry. 44, 2153–2162.
Privalov P.L., Potekhin S.A. 1986. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 131, 4–51.
Lyubarev A.E., Kurganov B.I. 2000. Analysis of irreversible protein denaturation by the method of differential scanning calorimetry. Usp. Biol. Khim. 40, 43–84.
Ladbury J. E., Doyle M. L. 2004. Biocalorimetry 2: Applications Calorimetry in the Biological Sciences. Chicherster: Wiley & Sons.
Frolova L., Le Goff X., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Justesen J., Kisselev L. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 372, 701–703.
Makhatadze G.I., Privalov P.L. 1990. Heat capacity of proteins: 1. Partial molar heat capacity of individual amino acid residues in aqueous solution: Hydration effect. J. Mol. Biol. 213, 375–384.
Prokop M., Damborsky J., Koca J. 2000. In silico construction of protein mutants and prediction of their activities. Bioinformatics. 16, 845–846.
Capriotti E., Fariselli P., Casadio R. 2004. A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 20, 163–168.
Chen Y.H., Yang J.T., Martinez H.M. 1972. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 11, 4120–4131.
Kononenko A.V., Dembo K.A., Kisselev L.L., Volkov V.V. 2004. Molecular morphology of eukaryotic class-1 translation termination factor eRF1 in solution. Mol. Biol. 38, 303–311.
Lyubarev A.E., Kurganov B.I. 1998. Modeling the process of irrevesible heat denaturation at variable temperature: 1. A model including two consecutive irreversible stages. Biokhimiya. 63, 434–440.
Kisselev L.L., Oparina N.Yu., Frolova L.Yu. 2000. Class-1 polypeptide chain release factors are structurally and functionally similar to suppressor tRNAs and comprise different structural-functional families of prokaryotic/mitochondrial and eukaryotic/archaebacterial factors. Mol. Biol. 34, 427–441.
Kolosov P., Frolova L., Seit-Nebi A., Dubovaya V., Kononenko A., Oparina N., Justesen J., Efimov A., Kisselev. L. 2005. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 33, 6418–6425.
Weng Z., Delisi C., Vajda S. 1997. Empirical free energy calculation: Comparison to calorimetric data. Protein Sci. 6, 1976–1984.
Author information
Authors and Affiliations
Additional information
Original Russian Text © V.A. Mitkevich, A.V. Kononenko, N.J. Oparina, P.M. Kolosov, A.A. Makarov, L.L. Kisselev, 2006, published in Molekulyarnaya Biologiya, 2006, Vol. 40, No. 1, pp. 100–110.
Rights and permissions
About this article
Cite this article
Mitkevich, V.A., Kononenko, A.V., Oparina, N.J. et al. Thermal denaturation of class 1 eukaryotic translation termination factor eRF1. Relationship between stability and functional activity of eRF1 mutants. Mol Biol 40, 86–95 (2006). https://doi.org/10.1134/S0026893306010134
Received:
Issue Date:
DOI: https://doi.org/10.1134/S0026893306010134