Abstract—
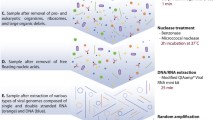
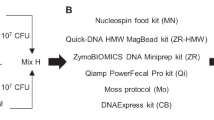
Metagenomics of the viromes associated with symbiotic microbial communities is a rapidly expanding field of research. Most of the relevant studies deal with the viral communities of human or animal feces. The novel technology of long-reads sequencing from Oxford Nanopore that became available during the last years provides outstanding new possibilities for research of uncultured viral communities. However, compared to classical NGS sequencing, this technology is much more sensitive to the quantity and quality of sequenced DNA. We developed a simple, cheap, and efficient protocol for extraction of total virome DNA from horse feces yielding the DNA samples suitable for long-read sequencing. This method can be adapted for the work with other difficult objects.



Similar content being viewed by others
REFERENCES
Babenko, V.V., Golomidova, A.K., Ivanov, P.A., Letarova, M.A., Kulikov, E.E., Manolov, A.I., Prokhorov, N.S., Kos-trukova, E.S., Matyushkina, D.M., Prilipov, A.G., Maslov, S., Belalov, I.S., Clokie, M.R.J.C., and Letarov, A.V., Phages associated with horses provide new insights into the dominance of lateral gene transfer in virulent bacteriophages evolution in natural systems, Biorxiv, 2019. https://doi.org/10.1101/542787
Castro-Mejia, J.L., Muhammed, M.K., Kot, W., Neve, H., Franz, C.M., Hansen, L.H., Vogensen, F.K., and Nielsen, D.S., Optimizing protocols for extraction of bacteriophages prior to metagenomic analyses of phage communities in the human gut, Microbiome, 2015, vol. 3, article 64. https://doi.org/10.1186/s40168-015-0131-4
Costa, M.C., Silva, G., Ramos, R.V., Staempfli, H.R., Arroyo, L.G., Kim, P., and Weese, J.S., Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses, Vet. J., 2015, vol. 205, pp. 74–80.
Jain, M., Olsen, H.E., Paten, B., and Akeson, M., The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community, Genome Biol., 2016, vol. 17, article 239.
Julliand, V. and Grimm, P., Horse Species Symposium: the microbiome of the horse hindgut: history and current knowledge1, J. Animal Sci., 2016, vol. 94, pp. 2262–2274.
Kulikov, E., Kropinski, A.M., Golomidova, A., Lingohr, E., Govorun, V., Serebryakova, M., Prokhorov, N., Letarova, M., Manykin, A., Strotskaya, A., and Letarov, A., Isolation and characterization of a novel indigenous intestinal N4-related coliphage vB_EcoP_G7C, Virology, 2012, vol. 426, pp. 93–99.
Milani, C., Casey, E., Lugli, G.A., Moore, R., Kaczorowska, J., Feehily, C., Mangifesta, M., Mancabelli, L., Duranti, S., Turroni, F., Bottacini, F., Mahony, J., Cotter, P.D., McAuliffe, F.M., van Sinderen, D., and Ventura, M., Tracing mother-infant transmission of bacteriophages by means of a novel analytical tool for shotgun metagenomic datasets: METAnnotatorX, Microbiome, 2018, vol. 6, article 145. https://doi.org/10.1186/s40168-018-0527-z
Shkoporov, A.N. and Hill, C., Bacteriophages of the human gut: the “known unknown” of the microbiome, Cell Host Microbe, 2019, vol. 25, pp. 195–209.
Tang, J.N., Zeng, Z.G., Wang, H.N., Yang, T., Zhang, P.J., Li, Y.L., Zhang, A.Y., Fan, W.Q., Zhang, Y., Yang, X., Zhao, S.J., Tian, G.B., and Zou, L.K., An effective method for isolation of DNA from pig faeces and comparison of five different methods, J. Microbiol. Methods, 2008, vol. 75, pp. 432–436.
Thomas, J.C., Khoury, R., Neeley, C.K., Akroush, A.M., and Davies, E.C., A fast CTAB method of human DNA isolation for polymerase chain reaction applications, Biochem. Educat., 1997, vol. 25, pp. 233–235.
Thurber, R.V., Haynes, M., Breitbart, M., Wegley, L., and Rohwer, F., Laboratory procedures to generate viral metagenomes, Nat. Protoc., 2009, vol. 4, pp. 470–483.
Vajda, B.P., Concentration and purification of viruses and bacteriophages with polyethylene glycol, Folia Microbiol., 1978, vol. 23, pp. 88–96.
Funding
This work was supported by the Russian Foundation for Basic Research, project no. 18-29-13029.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals or human subjects performed by any of the authors.
Additional information
Translated by D. Timchenko
Rights and permissions
About this article
Cite this article
Kulikov, E.E., Golomidova, A.K., Babenko, V.V. et al. A Simple Method for Extraction of the Horse Feces Virome DNA, Suitable for Oxford Nanopore Sequencing. Microbiology 89, 246–249 (2020). https://doi.org/10.1134/S002626172002006X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002626172002006X