Abstract
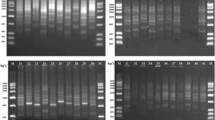
The presence of repeated DNA, viz. short tandemly repeated repetitive (STRR) and highly iterated palindrome (HIP) sequences was used as a typing technique for assessing genetic variability and phylogenetic relatedness of heterocystous cyanobacteria. Primers analogous to the STRR and HIP sequences were used to generate specific fingerprints for the twelve heterocystous cyanobacterial strains and a dendrogram was constructed. STRRmod and HIPTG primers revealed 100% polymorphism and yielded almost identical patterns. Anabaena sp. PCC 7120 clustered with Nostoc muscorum with both primers. Primer STRRmod supported the heterogeneity between Nostoc and Anabaena but HIPTG placed these two genera distinctly apart. STRRmod and HIPTG revealed that the members of the two orders were intermixed and thus suggesting a monophyletic origin of heterocystous cyanobacteria.
Similar content being viewed by others
References
Whitton, B.A. and Potts, M., Introduction of cyanobacteria, in The Ecology of Cyanobacteria: Their Diversity in Time and Space, Whitton, B.A. and Potts, M., Eds., Dordrecht Kluwer, 2000, pp. 1–10.
Castenholz, R.W., Oxygenic photosynthetic bacteria, in Bergey’s Manual of Systematic Bacteriology, 2nd ed., Boone, D.R. and Castenholz, R.W., Eds., New York: Springer, 2001, pp. 473–474.
Sigler, W.B., Bachofen, R., and Zeyer, J., Molecular characterization of endolithic cyanobacteria inhabiting exposed dolomite in central Switzerland, Environ. Microbiol., 2003, vol. 56, pp. 618–627.
Tandeau de Marsac, N. and Houmard, U., Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms, FEMS Microbiol. Rev., 1993, vol. 104, pp. 119–190.
Ferris, M.J., Kuhl, M., Wieland, A., and Ward, D.M., Cyanobacterial ecotypes in different optical microenvironments of a 68 degree C hot spring mat community revealed by 16S–23S rRNA internal transcribed spacer region variation, Appl. Environ. Microbiol., 2003, vol. 69, pp. 2893–2898.
Ward, D.M., Weller, R., and Bateson, M.M., 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community, Nature, 1990, vol. 345, pp. 63–65.
Otsuka, S., Suda, S., Li, R.H., Watanabe, M., Oyaizu, H., Matsumoto, S., and Watanabe, M.M., Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence, FEMS Microbiol. Lett., 1999, vol. 72, pp. 15–21.
Dyble, J., Paerl, H.W., and Neilan, B.A., Genetic characterization of Cylindrospermopsis raciborskii (Cyanobacteria) isolates from diverse geographic origins based on nifH and cpcBA IGS nucleotide sequence analysis, Appl. Environ. Microbiol., 2002, vol. 68, pp. 2567–2571.
Tsyrenova, D.D., Bryanskaya, A.V., Namsaraev, Z.B., and Akimov, V.N., Taxonomic and ecological characterization of cyanobacteria from some brackish and saline lakes of southern Transbaikal region, Microbiology (Moscow), 2011, vol. 80, pp. 216–227.
Mishra, A.K., Shukla, E., and Singh, S.S., Phylogenetic comparison among the heterocystous cyanobacteria based on a polyphasic approach, Protoplasma, 2013, vol. 250, pp. 77–94.
Singh, P., Singh, S.S., Elster, J., and Mishra, A.K., Molecular phylogeny, population genetics, and evolution of heterocystous cyanobacteria using nifH gene sequences, Protoplasma, 2013, vol. 250, pp. 751–764.
Jaspers, E. and Overmann, J., Ecological significance of microdiversity: identical 16s rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies, Appl. Environ. Microbiol., 2004, vol. 70, pp. 4831–4839.
Haverkamp, T., Acinas, S.G., Doeleman, M., Wollenzien, U., Huisman, J., and Stal, J., Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the Baltic Sea, The ISME J., 2009, vol. 3, pp. 397–408.
Robertson, B.R., Tezuka, N., and Watanabe, M.M., Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content, Int. J. Syst. Evol. Microbiol., 2001, vol. 51, pp. 861–871.
Mühling, M., Fuller, N.J., Millard, A., Somerfield, P.J., Marie, D., Wilson, W.H., Scanlan, D.J., Post, A.F., Joint, I., and Mann, N.H., Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton, Environ. Microbiol., 2005, vol. 7, pp. 499–508.
Zehr, J.P., Mellon, M.T., and Hiorns, W.D., Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages, Microbiology (UK), 1997, vol. 143, pp. 1443–1450.
Henson, B.J., Watson, L.E., and Barnum, S.R., Molecular differentiation of the heterocystous cyanobacteria, Nostoc and Anabaena, based on complete NifD sequences, Curr. Microbiol., 2002, vol. 45, pp. 161–164.
Versalovic, J., Koeuth, T., and Lupski, J.R., Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes, Nucl. Acids Res., 1991, vol. 19, pp. 6823–6831.
Smith, J.K., Parry, J.D., Day, J.G., and Smith, R.J., A PCR technique based on the Hip1 interspersed repetitive sequence distinguishes cyanobacterial species and strains, Microbiology (UK), 1998, vol. 144, pp. 2791–2801.
Rasmussen, U. and Svenning, M., Fingerprinting of cyanobacteria based on PCR with primers derived from short and long tandemly repeated repetitive sequences, Appl. Environ. Microbiol., 1998, vol. 64, pp. 265–272.
Lyra, C., Laamanen, M., Lehtimäki, J.M., Surakka, A., and Sivonen, K., Benthic cyanobacteria of the genus Nodularia are non toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia, Int. J. Syst. Evol. Microbiol., 2005, vol. 55, pp. 555–568.
Robinson, N.J., Robinson, P.J., Gupta, A., Bleasby, A.J., Whitton, B.A., and Morby, A.P., Singular overrepresentation of an octameric palindrome, Hip1, in DNA from many cyanobacteria, Nucl. Acids Res., 1995, vol. 23, pp. 729–735.
Selvakumar, G. and Gopalaswamy, G., PCR based fingerprinting of Westiellopsis cultures with short tandemly repeated repetitive (STRR) and highly iterated palindrome (HIP) sequences, Biologia, 2008, vol. 63, pp. 283–288.
Orcutt, K.M., Rasmussen, U., Webb, E.A., Waterbury, J.B., Gundersen, K., and Bergman, B., Characterization of Trichodesmium spp. by genetic techniques, Appl. Environ. Microbiol., 2002, vol. 68, pp. 2236–2245.
Valério, E., Chambel, L., Paulino, S., Faria, N., Pereira, P., and Tenreiro, R., Molecular identification, typing and traceability of cyanobacteria from freshwater reservoirs, Microbiology (UK), 2009, vol. 155, pp. 642–656.
Ezhilarasi, A. and Anand, N., Fingerprinting of repetitive DNA sequences in the genus Anabaena using PCR-based techniques, Afr. J. Microbiol. Res., 2010, vol. 4, pp. 590–598.
Rippka, R., Deruelles, J., Waterbury, J.B., Herdman, M., and Stanier, R.Y., Generic assignments, strain histories and properties of pure cultures of cyanobacteria, J. Gen. Microbiol., 1979, vol. 111, pp. 1–61.
Zheng, W.W., Nilsson, M., Bergman, B., and Rasmussen, U., Genetic diversity and classification of cyanobacteria in different Azolla species by the use of PCR fingerprinting, Theor. Appl. Genet., 1999, vol. 99, pp. 1187–1193.
Rohlf, F.J., Numerical Taxonomy and Multivariate Analysis System, version 1.80, Setauket, NY: Exeter Software, 1993.
Komárek, J. and Mareš, J., An update to modern taxonomy (2011) of freshwater planktic heterocystous cyanobacteria, Hydrobiology, 2012. doi: 10.1007/s10750-012-1027-y
Versalovic, J., Schneider, M., de Bruijn, F.J., and Lupski, J.R., Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR), Methods Cell Biol., 1994, vol. 5, pp. 25–40.
Katayama, T., Okamoto, S., Narikawa, R., Fujisawa, T., Kawashima, S., Itoh, M., Ohmori, M., and Kanehisa, M., Comprehensive analysis of tandem repeat sequences in cyanobacteria genome, Genome Informatics, 2002, vol. 13, pp. 400–401.
Mazel, D., Houmard, J., Castets, A.M., and de Marsac, N., Highly repetitive DNA sequences in cyanobacterial genomes, J. Bacteriol., 1990, vol. 172, pp. 2755–2761.
Rippka, R. and Herdman, M., Pasteur Culture Collection of Cyanobacterial Strains in Axenic Culture, Catalogue and Taxonomic Handbook, vol. 1, Paris: Institut Pasteur, 1992.
Shukla, E., Singh, S.S., Singh, P., and Mishra, A.K., Chemotaxonomy of heterocystous cyanobacteria using FAME profiling as species markers, Protoplasma, 2012, vol. 249, pp. 651–661.
Lyra, C., Suomalainen, S., Gugger, M., Vezie, C., Sundman, P., Paulin, L., and Sivonen, K., Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera, Int. J. Syst. Evol. Microbiol., 2001, vol. 51, pp. 513–526.
Svenning, M.M., Eriksson, T., and Rasmussen, U., Phylogeny of symbiotic cyanobacteria within the genus Nostoc based on 16S rDNA sequence analyses, Arch. Microbiol., 2005, vol. 183, pp. 19–26.
Halinen, K., Fewer, D.P., Shivonen, L.M., Lyra, C., Eronen, E., and Sivonen, K., Genetic diversity in strains of the genus Anabaena isolated from planktonic and benthic habitats of the Gulf of Finland (Baltic Sea), FEMS Microbiol. Ecol., 2008, vol. 64, pp. 199–208.
Fox, G.E., Wisotzkey, J.D., and Jurtshuk, P., How close is close: 16S ribosomal RNA sequence identity may not be sufficient to guarantee species identity, Int. J. Syst. Bacteriol., 1992, vol. 64, pp. 166–170.
Gugger, M. and Hoffmann, L., Polyphyly of the true branching cyanobacteria (Stigonematales), Int. J. Syst. Evol. Microbiol., 2004, vol. 54, pp. 349–357.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Shukla, E., Singh, S.S. & Mishr, A.K. Fingerprinting and phylogeny of some heterocystous cyanobacteria using short tandemly repeated repetitive and highly iterated palindrome sequences. Microbiology 82, 801–808 (2013). https://doi.org/10.1134/S0026261714010123
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261714010123