Abstract
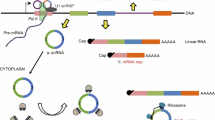
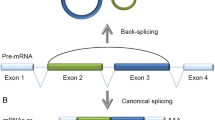
The roles that noncoding RNAs play in various cells have been a focus of intense research in recent years. Circular RNAs (circRNAs) are noncoding RNAs that were initially thought to be junk by-products of splicing. Many circRNAs have been found to demonstrate important regulatory functions and, in particular, to act as miRNA sponges. The function of an miRNA sponge was compared with the function of a classic transcription factor (TF)-dependent regulatory loop in terms of efficiency, working rate, and noise characteristics. A circRNA with multiple binding sites for miRNA was found to act more efficiently than a TF and the respective loop worked faster, but only when the binding sites were not fully saturated with miRNA molecules. The noise characteristics of the circRNA loop were significantly worse with an increasing number of binding sites. A circRNA with one binding site was shown to be inefficient as an miRNA sponge. The circRNA-mediated regulation was assumed to provide a specific tool to the cell.





Similar content being viewed by others
REFERENCES
J. Greene, A. M. Baird, L. Brady, et al., Front. Mol. Biosci. 4, 38 (2017).
S. Guil and M. Esteller, Trends Biochem. Sci. 40 (5), 248 (2015).
L. M. Holdt, A. Kohlmaier, and D. Teupser, Cell. Mol. Life Sci. 75, 1071 (2018).
J. Salzman, C. Gawad, P. L. Wang, et al., PLoS One 7 (2), e30733 (2012).
T. B. Hansen, J. Kjems, and C. K. Damgaard, Cancer Res. 73, 5609 (2013).
I. Jost, L. A. Shalamova, G. K. Gerresheim, et al., RNA Biol. 15 (8), 1032 (2018). https://doi.org/10.1080/15476286.2018.1435248
S. Memczak, M. Jens, A. Elefsinioti, et al., Nature 495, 333, (2013). https://doi.org/10.1038/nature11928
M. Wang, F. Yu, W. Wu, et al., Int. J. Biol. Sci. 13, 1497 (2017). https://doi.org/10.7150/ijbs.22531
R. Ashwal-Fluss, M. Meyer, N. R. Pamudurti, et al., Mol. Cell 56 (1), 55 (2014).
L. L. Chen and L. Yang, RNA Biol. 12 (4), 381 (2015).
Y. Dong, D. He, Z. Peng, et al., J. Hematol. Oncol. 10, Art. 2 (2017). https://doi.org/10.1186/s13045-016-0370-2
J. Singh and R. A. Padgett, Nat. Struct. Mol. Biol. 16 (11), 1128 (2009).
J. Hnilicova and D. Stanek, Nucleus 2 (3), 182 (2011).
L. Xu, H. Chen, X. Hu, et al., Mol. Biol. Evol. 23 (6), 1107 (2006).
Y. Taniguchi, P. J. Choi, G. W. Li, et al., Science 329 (5991), 533 (2010).
S. O. Olofsson, K. Bostrom, P. Carlsson, et al., Am. Heart J. 113 (2, Pt. 2), 446 (1987).
M. K. Doherty, D. E. Hammond, M. J. Clague, et al., J. Proteome Res. 8 (1), 104 (2009).
T. C. Chang and J. T. Mendell, Annu. Rev. Genomics Hum. Genet. 8, 215 (2007).
D. T. Gillespie, Phys. Rev. E 54 (2), 2084 (1996).
M. Osella, C. Bosia, D. Cora, and M. Caselle, PLoS Comput. Biol. 7 (3), e1001101 (2010).
S. Tej, K. Gaurav, and S. Mukherji, Phys. Biol. 16 (4), 046008 (2019). https://doi.org/10.1088/1478-3975/ab1563
M. A. Duk, M. G. Samsonova, and A. M. Samsonov, BMC Genomics 15 (Suppl. 12), S9 (2014). https://doi.org/10.1186/1471-2164-15-S12-S9
ACKNOWLEDGMENTS
We are grateful to S.A. Rukolaine for advice.
Funding
M.A. Duk acknowledges the support from research program no. 0040-2019-0003 Nonlinear Processes and Mechanisms of Mass Transfer in Condensed Media and Biostructures of the Ioffe Physical Technical Institute (study of the circRNA role). M.G. Samsonova acknowledges support from the Ministry of Education and Science of the Russian Federation (contract no. 075-15-2020-934 Advanced Digital Technologies with the St. Petersburg Polytechnic University) (verification of the biological significance of the study).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
This work does not contain any studies involving animals or human subjects performed by any of the authors.
Additional information
Translated by T. Tkacheva
Abbreviations: TF, transcription factor.
Rights and permissions
About this article
Cite this article
Duk, M.A., Samsonova, M.G. The Pros and Cons of Circular RNAs as miRNA Sponges. BIOPHYSICS 66, 8–16 (2021). https://doi.org/10.1134/S0006350921010036
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006350921010036