Abstract
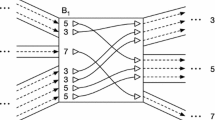
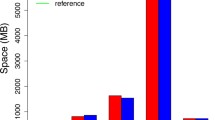
The problem of storage of the sequences of a number of closely related genomes and analysis of genome variations is considered. A genome graph with the structure of an acyclic directed graph is used to store matching sections of sequences and known variants. An algorithm for rapid mapping of reads to the genome graph is developed to align the individual nucleotide sequence fragments to the genome graph. The algorithm combines rapid searching using hash tables with the algorithm of dynamic programming and solves the problem of exponential growth in the number of paths on the graph. The implementation of the genome graph and the algorithm of the alignment of reads is developed. A comparison with the best-known programs with similar functionality is made.
Similar content being viewed by others
References
B. Paten, A. M. Novak, J. M. Eizenga, and E. Garrison, Genome Res. 27 (5), 665 (2017).
D. R. Zerbino and E. Birney, Genome Res. 18 (5), 821 (2008).
R. Chikhi and G. Rizk, Algorithms Mol. Biol. 8 (1), (2013).
R. Luo, B. Liu, Y. Xie, et al., GigaScience 1, (2012).
P. Compeau, P. A. Pevzner, and G. Tesler, Nature Biotechnol. 29 (11), 987 (2011).
A. Bankevich, S. Nurk, D. Antipov, et al., J. Comput. Biol. 19 (5), 455 (2012).
K. Bradnam, J. Fass, A. Alexandrov, et al., Giga-Science 2 (1), (2013).
A. M. Novak, E. Garrison, and B. Paten, Algorithms Mol. Biol. 12 (18), (2017).
S. Altschul, W. Gish, W. Miller, et al., J. Mol. Biol. 215 (3), 403 (1990).
H. Li and R. Durbin, Bioinformatics 25 (14), 1754 (2009).
P. Ferragina, F. Luccio, G. Manzini, et al., J. ACM 57 (1), 1 (2009).
J. Siren, N. Valimaki, and V. Makinen, IEEE/ACM Trans. Comput. Biol. Bioinform. 11 (2), 375 (2014).
J. Sirén, arXiv, 1604.06605 (2017).
GitHub: vg, 2014. https://github.com/vgteam/vg. Cited March 10, 2018.
GitHub: A Graph/Smith-Waterman (partial order) aligner/realigner, 2013. https://github.com/ekg/glia. Cited March 10, 2018.
NCBI: Escherichia coli IAI39 (E. coli) assembly, 2008. https://www.ncbi.nlm.nih.gov/assembly/GCF_000026345.1. Cited February 8, 2018.
NCBI: Escherichia coli str. K-12 substr. MG1655 (E. coli) assembly, 2013. https://www.ncbi.nlm.nih.gov/assembly/GCF_000005845.2. Cited February 8, 2018.
A. Darling, B. Mau, F. Blattner, et al., Genome Res. 14 (7), 1394 (2004).
K. Katoh and D. Standley, Mol. Biol. Evol. 30 (4), 772 (2013).
J. Ju, D. Kim, L. Bi, et al., Proc. Natl Acad. Sci. U. S. A. 103 (52), 19635 (2006).
B. Langmead and S. Salzberg, Nat. Methods 9 (4), 357 (2012).
D. Kim, B. Langmead, and S. Salzberg, Nat. Methods 12 (4), 357 (2015).
Samtools Organisation and Repositories: The Variant Call Format (VCF) Version 4.2 Specification, 2017. https://samtools.github.io/hts-specs/VCFv4.2.pdf. Cited March 20, 2018.
M. View, H. Olsen, B. Paten, et al., Genome Biol. 17 (1), 239 (2016).
H. Buermans and J. den Dunnen, Biochim. Biophys. Acta 1842 (10), 1932 (2014).
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © S.N. Petrov, L.A. Uroshlev, A.S. Kasyanov, V.Yu. Makeev, 2018, published in Biofizika, 2018, Vol. 63, No. 3, pp. 421–429.
Rights and permissions
About this article
Cite this article
Petrov, S.N., Uroshlev, L.A., Kasyanov, A.S. et al. An Efficient Algorithm for Mapping of Reads to a Genome Graph Using an Index Based on Hash Tables and Dynamic Programming. BIOPHYSICS 63, 311–317 (2018). https://doi.org/10.1134/S0006350918030193
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006350918030193