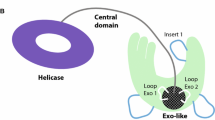
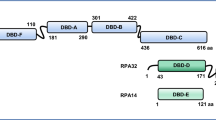
Abstract
Transmission of genetic information depends on successful completion of DNA replication. Genomic DNA is subjected to damage on a daily basis. DNA lesions create obstacles for DNA polymerases and can lead to the replication blockage, formation of DNA breaks, cell cycle arrest, and apoptosis. Cells have evolutionary adapted to DNA damage by developing mechanisms allowing elimination of lesions prior to DNA replication (DNA repair) and helping to bypass lesions during DNA synthesis (DNA damage tolerance). The second group of mechanisms includes the restart of DNA synthesis at the sites of DNA damage by DNA primase-polymerase PrimPol. Human PrimPol was described in 2013. The properties and functions of this enzyme have been extensively studied in recent years, but very little is known about the regulation of PrimPol and association between the enzyme dysfunction and diseases. In this review, we described the mechanisms of human PrimPol regulation in the context of DNA replication, discussed in detail interactions of PrimPol with other proteins, and proposed possible pathways for the regulation of human PrimPol activity. The article also addresses the association of PrimPol dysfunction with human diseases.



Similar content being viewed by others
References
Iyer, L. M., Koonin, E. V., Leipe, D. D., and Aravind, L. (2005) Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members, Nucleic Acids Res., 33, 3875-3896, https://doi.org/10.1093/nar/gki702.
García-Gómez, S., Reyes, A., Martínez-Jiménez, M. I., Chocrón, S., Mourón, S., Terrados, G., Powell, C., Salido, E., Méndez, J., Holt, I. J., and Blanco, L. (2013) PrimPol, an archaic primase/polymerase operating in human cells, Mol. Cell, 52, 541-553, https://doi.org/10.1016/j.molcel.2013.09.025.
Wan, L., Lou, J., Xia, Y., Su, B., Liu, T., Cui, J., Sun, Y., Lou, H., and Huang, J. (2013) HPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity, EMBO Rep., 14, 1104-1112, https://doi.org/10.1038/embor.2013.159.
Bianchi, J., Rudd, S. G., Jozwiakowski, S. K., Bailey, L. J., Soura, V., Taylor, E., Stevanovic, I., Green, A. J., Stracker, T. H., Lindsay, H. D., and Doherty, A. J. (2013) Primpol bypasses UV photoproducts during eukaryotic chromosomal DNA replication, Mol. Cell, 52, 566-573, https://doi.org/10.1016/j.molcel.2013.10.035.
Kobayashi, K., Guilliam, T. A., Tsuda, M., Yamamoto, J., Bailey, L. J., Iwai, S., Takeda, S., Doherty, A. J., and Hirota, K. (2016) Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleoside, Cell Cycle, 15, 1997-2008, https://doi.org/10.1080/15384101.2016.1191711.
Bailey, L. J., Bianchi, J., Hégarat, N., Hochegger, H., and Doherty, A. J. (2016) PrimPol-deficient cells exhibit a pronounced G2 checkpoint response following UV damage, Cell Cycle, 15, 908-918, https://doi.org/10.1080/15384101.2015.1128597.
Bailey, L. J., Bianchi, J., and Doherty, A. J. (2019) PrimPol is required for the maintenance of efficient nuclear and mitochondrial DNA replication in human cells, Nucleic Acids Res., 47, 4026-4038, https://doi.org/10.1093/nar/gkz056.
Makarova, A. V., Boldinova, E. O., Belousova, E. A., and Lavrik, O. I. (2018) In vitro lesion bypass by human PrimPol, DNA Repair (Amst.), 70, 18-24, https://doi.org/10.1016/j.dnarep.2018.07.009.
Keen, B. A., Jozwiakowski, S. K., Bailey, L. J., Bianchi, J., and Doherty, A. J. (2014) Molecular dissection of the domain architecture and catalytic activities of human PrimPol, Nucleic Acids Res., 42, 5830-5845, https://doi.org/10.1093/nar/gku214.
Stojkovic, G., Makarova, A. V., Wanrooij, P. H., Forslund, J., Burgers, P. M. J., and Wanrooij, S. (2016) Oxidative DNA damage stalls the human mitochondrial replisome, Sci. Rep., 6, 28942, https://doi.org/10.1038/srep28942.
Zafar, M. K., Ketkar, A., Lodeiro, M. F., Cameron, C. E., and Eoff, R. L. (2014) Kinetic analysis of human PrimPol DNA polymerase activity reveals a generally error-prone enzyme capable of accurately bypassing 7,8-dihydro-8-oxo-2′-deoxyguanosine, Biochemistry, 53, 6584-6594, https://doi.org/10.1021/bi501024u.
Martínez-Jiménez, M. I., García-Gómez, S., Bebenek, K., Sastre-Moreno, G., Calvo, P. A., Díaz-Talavera, A., Kunkel, T. A., and Blanco, L. (2015) Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol, DNA Repair (Amst.), 29, 127-138, https://doi.org/10.1016/j.dnarep.2015.02.013.
Boldinova, E. O., Yudkina, A. V., Shilkin, E. S., Gagarinskaya, D. I., Baranovskiy, A. G., Tahirov, T. H., Zharkov, D. O., and Makarova, A. V. (2021) Translesion activity of PrimPol on DNA with cisplatin and DNA-protein cross-links, Sci. Rep., 11, 17588, https://doi.org/10.1038/s41598-021-96692-y.
Boldinova, E. O., Ghodke, P. P., Sudhakar, S., Mishra, V. K., Manukyan, A. A., Miropolskaya, N., Pradeepkumar, P. I., and Makarova, A. V. (2022) Translesion synthesis across the N2-Ethyl-deoxyguanosine adduct by human PrimPol, ACS Chem. Biol., 17, 3238-3250, https://doi.org/10.1021/acschembio.2c00717.
Guilliam, T. A., Brissett, N. C., Ehlinger, A., Keen, B. A., Kolesar, P., Taylor, E., Bailey, L. J., Lindsay, H. D., Chazin, W. J., and Doherty, A. J. (2017) Molecular basis for PrimPol recruitment to replication forks by RPA, Nat. Commun., 8, 15222, https://doi.org/10.1038/ncomms15222.
Quinet, A., Tirman, S., Jackson, J., Šviković, S., Lemaçon, D., Carvajal-Maldonado, D., González-Acosta, D., Vessoni, A. T., Cybulla, E., Wood, M., Tavis, S., Batista, l. F. Z., Méndez, J., Sale, J. S., and Vindigni, A. (2019) PRIMPOL-mediated adaptive response suppresses replication fork Reversal in BRCA-deficient cells, Mol. Cell, 77, 461-474.e9, https://doi.org/10.1016/j.molcel.2019.10.008.
Piberger, A. L., Bowry, A., Kelly, R., Walker, A. K., Gonzalez, D., Bailey, L. J., Doherty, A. J., Méndez, J., Morris, J. R., Bryant, H. E., and Petermann, E. (2020) PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts, Nat. Commun., 11, 5863, https://doi.org/10.1038/s41467-020-19570-7.
González‐Acosta, D., Blanco‐Romero, E., Ubieto‐Capella, P., Mutreja, K., Míguez, S., Llanos, S., García, F., Muñoz, J., Blanco, L., Lopes, M., and Méndez, J. (2021) PrimPol‐mediated repriming facilitates replication traverse of DNA interstrand crosslinks, EMBO J., 40, e106355, https://doi.org/10.15252/embj.2020106355.
Schiavone, D., Jozwiakowski, S. K., Romanello, M., Guilbaud, G., Guilliam, T. A., Bailey, L. J., Sale, J. E., and Doherty, A. J. (2016) PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells, Mol. Cell, 61, 161-169, https://doi.org/10.1016/j.molcel.2015.10.038.
Zhao, F., Wu, J., Xue, A., Su, Y., Wang, X., Lu, X., Zhou, Z., Qu, J., and Zhou, X. (2013) Exome sequencing reveals CCDC111 mutation associated with high myopia, Hum. Genet., 132, 913-921, https://doi.org/10.1007/s00439-013-1303-6.
Keen, B. A., Bailey, L. J., Jozwiakowski, S. K., and Doherty, A. J. (2014) Human PrimPol mutation associated with high myopia has a DNA replication defect, Nucleic Acids Res., 42, 12102-12111, https://doi.org/10.1093/nar/gku879.
Kasamo, K., Nakamura, M., Daimou, Y., and Sano, A. (2020) A PRIMPOL mutation and variants in multiple genes may contribute to phenotypes in a familial case with chronic progressive external ophthalmoplegia symptoms, Neurosci. Res., 157, 58-63, https://doi.org/10.1016/j.neures.2019.07.006.
Duong, V. N., Zhou, L., Martínez-Jiménez, M. I., He, L., Cosme, M., Blanco, L., Paintsil, E., Anderson, K. S. (2020) Identifying the role of PrimPol in TDF-induced toxicity and implications of its loss of function mutation in an HIV+ patient, Sci. Rep., 10, 9343, https://doi.org/10.1038/s41598-020-66153-z.
Díaz-Talavera, A., Calvo, P. A., González-Acosta, D., Díaz, M., Sastre-Moreno, G., Blanco-Franco, L., Guerra, S., Martínez-Jiménez, M. I., Méndez, J., and Blanco, L. (2019) A cancer-associated point mutation disables the steric gate of human PrimPol, Sci. Rep., 9, 1121, https://doi.org/10.1038/s41598-018-37439-0.
Picher, Á. J., Budeus, B., Wafzig, O., Krüger, C., García-Gómez, S., Martínez-Jiménez, M. I., Díaz-Talavera, A., Weber, D., Blanco, L., and Schneider, A. (2016) TruePrime is a novel method for whole-genome amplification from single cells based on TthPrimPol, Nat. Commun., 7, 13296, https://doi.org/10.1038/ncomms13296.
Agudo, R., Calvo, P. A., Martínez-Jiménez, M. I., and Blanco, L. (2017) Engineering human PrimPol into an efficient RNA-dependent-DNA primase/polymerase, Nucleic Acids Res., 45, 9046-9058, https://doi.org/10.1093/nar/gkx633.
Boldinova, E. O., Wanrooij, P. H., Shilkin, E. S., Wanrooij, S., and Makarova, A. V. (2017) DNA damage tolerance by eukaryotic DNA polymerase and primase PrimpPol, Int. J. Mol. Sci., 18, 1584, https://doi.org/10.3390/ijms18071584.
Tirman, S., Cybulla, E., Quinet, A., Meroni, A., and Vindigni, A. (2020) PRIMPOL ready, set, reprime! Crit. Rev. Biochem. Mol. Biol., 56, 17-30, https://doi.org/10.1080/10409238.2020.1841089.
Baretic, D., Jenkyn-bedford, M., Aria, V., Cannone, G., Skehel, M., and Yeeles, J. T. P. (2020) Cryo-EM structure of the fork protection complex bound to CMG at a replication fork, Mol. Cell, 78, 926-940.e13, https://doi.org/10.1016/j.molcel.2020.04.012.
Jones, M. L., Baris, Y., Taylor, M. R. G., and Yeeles, J. T. P. (2021) Structure of a human replisome shows the organisation and interactions of a DNA replication machine, EMBO J., 40, e108819, https://doi.org/10.15252/embj.2021108819.
Yuan, Z., Georgescu, R., Luna, R. De, Santos, A., Zhang, D., Bai, L., Yao, N. Y., Zhao, G., O’Donnell, M. E., and Li, H. (2019) Ctf4 organizes sister replisomes and Pol a into a replication factory, eLife, 8, e47405, https://doi.org/10.7554/eLife.47405.
Rzechorzek, N. J., Hardwick, S. W., Jatikusumo, V. A., Chirgadze, D. Y., and Pellegrini, L. (2020) CryoEM structures of human CMG – ATP-S – DNA and CMG – AND-1 complexes, Nucleic Acids Res., 48, 6980-6995, https://doi.org/10.1093/nar/gkaa429.
Kapadia, N., El-Hajj, Z. W., Zheng, H., Beattie, T. R., Yu, A., and Reyes-Lamothe, R. (2020) Processive activity of replicative DNA polymerases in the replisome of live eukaryotic cells, Mol. Cell, 80, 114-126.e8, https://doi.org/10.1016/j.molcel.2020.08.014.
Lewis, J. S., Spenkelink, L. M., Schauer, G. D., Yurieva, O., Mueller, S. H., Natarajan, V., Kaur, G., Maher, C., Kay, C., O’Donnell, M. E., and van Oijen, A. M. (2020) Tunability of DNA polymerase stability during eukaryotic DNA replication, Mol. Cell, 77, 17-25.e5, https://doi.org/10.1016/j.molcel.2019.10.005.
Donnell, M. O., and Li, H. (2016) The eukaryotic replisome goes under the microscope, Curr. Biol., 26, R247-R256, https://doi.org/10.1016/j.cub.2016.02.034.
Bleichert, F., Botchan, M. R., and Berger, J. M. (2017) Mechanisms for initiating cellular DNA replication, Science, 355, eaah6317, https://doi.org/10.1126/science.aah6317.
Remus, D., Beuron, F., Tolun, G., Griffith, J. D., Morris, E. P., and Diffley, J. F. X. (2009) Concerted loading of Mcm2 – 7 double hexamers around DNA during DNA replication origin licensing, Cell, 139, 719-730, https://doi.org/10.1016/j.cell.2009.10.01.
Tanaka, S., and Araki, H. (2013) Helicase activation and establishment of replication forks at chromosomal origins of replication, Cold Spring Harb. Perspect. Biol., 5, a010371, https://doi.org/10.1101/cshperspect.a010371.
Perez-Arnaiz, P., Bruck, I., and Kaplan, D. L. (2016) Mcm10 coordinates the timely assembly and activation of the replication fork helicase, Nucleic Acids Res., 44, 315-329, https://doi.org/10.1093/nar/gkv1260.
Kim, C., Paulus, B. F., and Wold, M. S. (1994) Interactions of human replication protein A with oligonucleotides, Biochemistry, 33, 14197-14206, https://doi.org/10.1021/bi00251a031.
Yates, L. A., Aramayo, R. J., Pokhrel, N., Caldwell, C. C., Kaplan, J. A., Perera, R. L., Spies, M., Antony, E., and Zhang, X. (2018) A structural and dynamic model for the assembly of replication protein A on single-stranded DNA, Nat. Commun., 9, 5447, https://doi.org/10.1038/s41467-018-07883-7.
Pestryakov, P. E., and Lavrik, O. I. (2008) Mechanisms of single-stranded DNA-binding protein functioning in cellular DNA metabolism, Biochemistry (Moscow), 73, 1388-1404, https://doi.org/10.1134/S0006297908130026.
Bhat, K. P., and Cortez, D. (2018) RPA and RAD51: fork reversal, fork protection, and genome stability, Nat. Struct. Mol. Biol., 25, 446-453, https://doi.org/10.1038/s41594-018-0075-z.
Zou, Y., Liu, Y., Wu, X., and Shell, S. M. (2006) Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses, J. Cell Physiol., 208, 267-273, https://doi.org/10.1002/jcp.20622.
Jain, R., Aggarwal, A. K., and Rechkoblit, O. (2018) Eukaryotic DNA polymerases, Curr. Opin. Struct. Biol., 53, 77-87, https://doi.org/10.1016/j.sbi.2018.06.003.
Baranovskiy, A. G., Babayeva, N. D., Zhang, Y., Gu, J., Suwa, Y., Pavlov, Y. I., and Tahirov, T. H. (2016) Mechanism of concerted RNA–DNA primer synthesis by the human primosome, J. Biol. Chem., 291, 10006-10020, https://doi.org/10.1074/jbc.M116.717405.
Goswami, P., Abid Ali, F., Douglas, M. E., Locke, J., Purkiss, A., Janska, A., Eickhoff, P., Early, A., Nans, A., Cheung, A. M. C., Diffley, J. F. X., and Costa, A. (2018) Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome, Nat. Commun., 9, 5061, https://doi.org/10.1038/s41467-018-07417-1.
Guilliam, T. A., and Yeeles, J. T. (2021) The eukaryotic replisome tolerates leading‐strand base damage by replicase switching, EMBO J., 40, e107037, https://doi.org/10.15252/embj.2020107037.
Miyabe, I., Mizuno, K., Keszthelyi, A., Daigaku, Y., Skouteri, M., Mohebi, S., Kunkel, T. A., Murray, J. M., and Carr, A. M. (2015) Polymerase d replicates both strands after homologous recombination-dependent fork restart, Nat. Struct. Mol. Biol., 22, 932-938, https://doi.org/10.1038/nsmb.3100.
Zhou, Z. -X., Lujan, S. A., Burkholder, A. B., Charles, J. S., Dahl, J., Farrell, C. E., Williams, J. S., and Kunkel, T. A. (2021) How asymmetric DNA replication achieves symmetrical fidelity, Nat. Struct. Mol. Biol., 28, 1020-1028, https://doi.org/10.1038/s41594-021-00691-6.
Bulock, C. R., Xing, X., and Shcherbakova, P. V. (2020) DNA polymerase δ proofreads errors made by DNA polymerase e, Proc. Natl. Acad. Sci. USA, 117, 6035-6041, https://doi.org/10.1073/pnas.1917624117.
Sun, H., Ma, L., Tsai, Y., Abeywardana, T., Shen, B., and Zheng, L. (2022) Okazaki fragment maturation: DNA flap dynamics for cell proliferation and survival, Trends Cell Biol., 33, 221-234, https://doi.org/10.1016/j.tcb.2022.06.014.
Stodola, J. L., and Burgers, P. M. J. (2017) Mechanism of lagging-strand DNA replication in eukaryotes, Adv. Exp. Med. Biol., 1042, 117-133, https://doi.org/10.1007/978-981-10-6955-0-6.
Sobhy, M. A., Tehseen, M., Takahashi, M., Bralić, A., De Biasio, A., and Hamdan, S. M. (2021) Implementing fluorescence enhancement, quenching, and FRET for investigating flap endonuclease 1 enzymatic reaction at the single-molecule level, Comput. Struct. Biotechnol. J., 19, 4456-4471, https://doi.org/10.1016/j.csbj.2021.07.029.
Liu, S., Lu, G., Ali, S., Liu, W., Zheng, L., Dai, H., Li, H., Xu, H., Hua, Y., Zhou, Y., Ortega, J., Li, G., Kunkel, T. A., and Shen, B. (2015) Okazaki fragment maturation involves a-segment error editing by the mammalian FEN 1/MutSa functional complex, EMBO J., 34, 1829-1843, https://doi.org/10.15252/embj.201489865.
Howes, T. R. L., and Tomkinson, A. E. (2012) DNA ligase I, the replicative DNA ligase, The Eukaryotic Replisome: a Guide to Protein Structure and Function (MacNeill, S., ed.) Dordrecht, Springer, pp. 327-341, https://doi.org/10.1007/978-94-007-4572-8.
Sverzhinsky, A., Tomkinson, A. E., and Pascal, J. M. (2022) Cryo-EM structures and biochemical insights into heterotrimeric PCNA regulation of DNA ligase, Structure, 30, 371-385.e5, https://doi.org/10.1016/j.str.2021.11.002.
Gonzalez-Magana, A., and Blanco, F. J. (2020) Human PCNA structure, function, and interactions, Biomolecules, 10, 570, https://doi.org/10.3390/biom10040570.
Stokes, K., Winczura, A., Song, B., De Piccoli, G., and Grabarczyk, D. B. (2020) Ctf18-RFC and DNA Pol epsilon form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication, Nucleic Acids Res., 48, 8128-8145, https://doi.org/10.1093/nar/gkaa541.
Baris, Y., Taylor, M. R. G., Aria, V., and Yeeles, J. T. P. (2022) Fast and efficient DNA replication with purified human proteins, Nature, 606, 204-210, https://doi.org/10.1038/s41586-022-04759-1.
Chilkova, O., Stenlund, P., Isoz, I., Stith, C. M., Burgers, P. M., and Johansson, E. (2007) The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA, Nucleic Acids Res., 35, 6588-6597, https://doi.org/10.1093/nar/gkm741.
Lancey, C., Tehseen, M., Raducanu, V., Rashid, F., Merino, N., Ragan, T. J., Savva, C. G., Zaher, M. Z., Shirbini, A., Blanco, F. J., Hamdan, S. M., and De Biasio, A. (2020) Structure of the processive human Pol δ holoenzyme, Nat. Commun., 11, 1109, https://doi.org/10.1038/s41467-020-14898-6.
Craggs, T. D., Hutton, R. D., Brenlla, A., White, M. F., and Penedo, J. C. (2014) Single-molecule characterization of Fen1 and Fen1/PCNA complexes acting on flap substrates, Nucleic Acids Res., 42, 1857-1872, https://doi.org/10.1093/nar/gkt1116.
Smits, V. A. J., Cabrera, E., Freire, R., and Gillespie, D. A. (2019) Claspin – checkpoint adaptor and DNA replication factor, EMBO J., 286, 441-455, https://doi.org/10.1111/febs.14594.
Hsiao, H. W., Chun, C., and Hisao, Y. (2021) Roles of Claspin in regulation of DNA replication, replication stress responses and oncogenesis in human cells, Genome Instab. Dis., 2, 263-280, https://doi.org/10.1007/s42764-021-00049-8.
Bertoli, C., Skotheim, J. M., De Bruin, R. A. M., and Street, G. (2013) Control of cell cycle transcription during G1 and S phases, Nat. Rev. Mol. Cell Biol., 14, 518-528, https://doi.org/10.1038/nrm3629.
Hume, S., Dianov, G. L., and Ramadan, K. (2020) A unified model for the G1/S cell cycle transition, Nucleic Acids Res., 48, 12483-12501, https://doi.org/10.1093/nar/gkaa1002.
Malumbres, M., and Barbacid, M. (2009) Cell cycle, CDKs and cancer: a changing paradigm, Nat. Rev. Cancer, 9, 153-166, https://doi.org/10.1038/nrc2602.
Mujwar, S., Mojzych, M., and Kontek, R. (2022) Cyclin-dependent kinases in DNA damage response, Biochim. Biophy. Acta Rev. Cancer, 1877, 188716, https://doi.org/10.1016/j.bbcan.2022.188716.
Williams, A. B., and Schumacher, B. (2016) p53 in the DNA-damage-repair process, Cold Spring Harb. Perspect. Med., 6, a026070, https://doi.org/10.1101/cshperspect.a026070.
Barnum, K. J., and Connell, M. J. O. (2014) Cell cycle regulation by checkpoints, Methods Mol. Biol., 1170, 29-40, https://doi.org/10.1007/978-1-4939-0888-2.
Patil, M., Pabla, N., and Dong, Z. (2013) Checkpoint kinase 1 in DNA damage response and cell cycle regulation, Cell. Mol. Life Sci., 70, 4009-4021, https://doi.org/10.1007/s00018-013-1307-3.
Taylor, M. R. G., and Yeeles, J. T. P. (2019) Dynamics of replication fork progression following helicase – polymerase uncoupling in eukaryotes, J. Mol. Biol., 431, 2040-2049, https://doi.org/10.1016/j.jmb.2019.03.011.
Byun, T. S., Pacek, M., Yee, M., Walter, J. C., and Cimprich, K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint, Genes Dev., 19, 1040-1052, https://doi.org/10.1101/gad.1301205.
Wong, R. P., Garcia-Rodriguez, N., Zilio, N., Hanulova, M., and Ulrich, H. (2020) Processing of DNA polymerase-blocking lesions during genome replication is spatially and temporally segregated from replication forks, Mol. Cell, 77, 3-16.e4, https://doi.org/10.1016/j.molcel.2019.09.015.
Zou, L., and Elledge, S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes, Science, 300, 1542-1548, https://doi.org/10.1126/science.1083430.
Blackford, A. N., and Jackson, S. P. (2017) ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response, Mol. Cell, 66, 801-817, https://doi.org/10.1016/j.molcel.2017.05.015.
Saldivar, J. C., Cortez, D., and Cimprich, K. A. (2017) The essential kinase ATR: ensuring faithful duplication of a challenging genome, Nat. Rev. Mol. Cell Biol., 18, 622-636, https://doi.org/10.1038/nrm.2017.67.
Cimprich, K. A., and Cortez, D. (2008) ATR: an essential regulator of genome integrity, Nat. Rev. Mol. Cell Biol., 9, 616-627, https://doi.org/10.1038/nrm2450.
Li, X., and Heyer, W.-D. (2008) Homologous recombination in DNA repair and DNA damage tolerance, Cell Res., 18, 99-113, https://doi.org/10.1038/cr.2008.1.
Chang, D. J., and Cimprich, K. A. (2009) DNA damage tolerance: when it’s OK to make mistakes, Nat. Chem. Biol., 5, 82-90, https://doi.org/10.1038/nchembio.139.
Neelsen, K. J., and Lopes, M. (2015) Replication fork reversal in eukaryotes: from dead end to dynamic response, Nat. Rev. Mol. Cell Biol., 16, 207-220, https://doi.org/10.1038/nrm3935.
Berti, M., Cortez, D., and Lopes, M. (2020) The plasticity of DNA replication forks in response to clinically relevant genotoxic stress, Nat. Rev. Mol. Cell Biol., 21, 633-665, https://doi.org/10.1038/s41580-020-0257-5.
Calvo, P. A., Sastre-Moreno, G., Perpiñá, C., Guerra, S., Martínez-Jiménez, M. I., and Blanco, L. (2019) The invariant glutamate of human PrimPol DxE motif is critical for its Mn2+-dependent distinctive activities, DNA Repair (Amst.), 77, 65-75, https://doi.org/10.1016/j.dnarep.2019.03.006.
Boldinova, E. O., Manukyan, A. A., and Makarova, A. V. (2021) The DNA ligands Arg47 and Arg76 are crucial for catalysis by human PrimPol, DNA Repair (Amst.), 100, 103048, https://doi.org/10.1016/j.dnarep.2021.103048.
Mourón, S., Rodriguez-Acebes, S., Martínez-Jiménez, M. I., García-Gómez, S., Chocrón, S., Blanco, L., and Méndez, J. (2013) Repriming of DNA synthesis at stalled replication forks by human PrimPol, Nat. Struct. Mol. Biol., 20, 1383-1389, https://doi.org/10.1038/nsmb.2719.
Martínez-Jiménez, M. I., Calvo, P. A., García-Gómez, S., Guerra-González, S., and Blanco, L. (2018) The Zn-finger domain of human PrimPol is required to stabilize the initiating nucleotide during DNA priming, Nucleic Acids Res., 46, 4138-4151, https://doi.org/10.1093/nar/gky230.
Tokarsky, E. J., Wallenmeyer, P. C., Phi, K. K., and Suo, Z. (2016) Significant impact of divalent metal ions on the fidelity, sugar selectivity, and drug incorporation efficiency of human PrimPol, DNA Repair (Amst.), 49, 51-59, https://doi.org/10.1016/j.dnarep.2016.11.003.
Traut, T. W. (1994) Physiological concentrations of purines and pyrimidines, Mol. Cel. Biochem., 140, 1-22, https://doi.org/10.1007/BF00928361.
McElhinny, S. A., Watts, B. E., Kumar, D., Watt, D. L., and Lundström, E. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases, Proc. Natl. Acad. Sci. USA, 107, 4949-4954, https://doi.org/10.1073/pnas.0914857107.
Blanco, L., Calvo, P. A., Diaz-Talavera, A., Carvalho, G., Calero, N., Martínez-Carrón, A., Velázquez-Ruiz, C., Villadangos, S., Guerra, S., and Martínez-Jiménez, M. I. (2019) Mechanism of DNA primer synthesis by human PrimPol, Enzymes, 45, 289-310, https://doi.org/10.1016/bs.enz.2019.06.003.
Butler, T. J., Estep, K. N., Sommers, J. A., Maul, R. W., Moore, A. Z., Bandinelli, S., Cucca, F., Tuke, M. A., Wood, A. R., Bharti, S. K., Bogenhagen, D. F., Yakubovskaya, E., Garcia-Diaz, M., Guilliam, T. A., Byrd, A. K., Raney, K. D., Doherty, A. J., Ferrucci, L., Schlessinger, D., Ding, J., and Brosh, R. M., Jr. (2020) Mitochondrial genetic variation is enriched in G-quadruplex regions that stall DNA synthesis in vitro, Hum. Mol. Genet., 29, 1292-1309, https://doi.org/10.1093/hmg/ddaa043.
Boldinova, E. O., Belousova, E. A., Gagarinskaya, D. I., Maltseva, E. A., Khodyreva, S. N., Lavrik, O. I., and Makarova, A. V. (2020) Strand displacement activity of PrimPol, Int. J. Mol. Sci., 21, 9027, https://doi.org/10.3390/ijms21239027.
Torregrosa-Muñumer, R., Forslund, J., Goffart, S., Pfeiffer, A., Stojkovic, G., Carvalho, G., Al-Furoukh, N., Blanco, L., Wanrooij, S., and Pohjoismäki, J. L. O. (2017) PrimPol is required for replication reinitiation after mtDNA damage, Proc. Natl. Acad. Sci. USA, 114, 11398-11403, https://doi.org/10.1073/pnas.1705367114.
Mehta, K. P. M., Thada, V., Zhao, R., Krishnamoorthy, A., Leser, M., Rose, K. L., and Cortez, D. (2022) CHK1 phosphorylates PRIMPOL to promote replication stress tolerance, Sci. Adv., 8, eabm0314, https://doi.org/10.1126/sciadv.abm0314.
Bailey, L. J., Teague, R., Kolesar, P., Bainbridge, L. J., Lindsay, H. D., and Doherty, A. J. (2021) PLK1 regulates the PrimPol damage tolerance pathway during the cell cycle, Sci. Adv., 7, eabh1004, https://doi.org/10.1126/sciadv.abh1004.
Keiji, T. (2009) The proteasome: overview of structure and functions, Proc. Jpn., 85, 12-36, https://doi.org/10.2183/pjab/85.12.
Yan, Y., Xu, Z., Huang, J., Guo, G., Gao, M., Kim, W., Zeng, X., Kloeber, J. A., Zhu, Q., Zhao, F., Luo, K., and Lou, Z. (2020) The deubiquitinase USP36 Regulates DNA replication stress and confers therapeutic resistance through PrimPol stabilization, Nucleic Acids Res., 48, 12711-12726, https://doi.org/10.1093/nar/gkaa1090.
Yoshimura, A., Oikawa, M., Jinbo, H., Hasegawa, Y., Enomoto, T., and Seki, M. (2019) WRNIP1 controls the amount of PrimPol, Biol. Pharm. Bull., 42, 764-769, https://doi.org/10.1248/bpb.b18-00955.
Guilliam, T. A., Jozwiakowski, S. K., Ehlinger, A., Barnes, R. P., Rudd, S. G., Bailey, L. J., Skehel, J. M., Eckert, K. A., Chazin, W. J., and Doherty, A. J. (2015) Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins, Nucleic Acids Res., 43, 1056-1068, https://doi.org/10.1093/nar/gku1321.
Martínez-Jiménez, M. I., Lahera, A., and Blanco, L. (2017) Human PrimPol activity is enhanced by RPA, Sci. Rep., 7, 783, https://doi.org/10.1038/s41598-017-00958-3.
Boldinova, E. O., Baranovskiy, A. G., Gagarinskaya, D. I., Manukyan, A. A., Makarova, A. V., and Tahirov, T. H. (2023) The role of catalytic and regulatory domains of human PrimPol in DNA binding and synthesis, Nucleic Acids Res., gkad507, https://doi.org/10.1093/nar/gkad507.
Gagarinskaya, D. I., and Makarova, A. V. (2020) A multifunctional protein PolDIP2 in DNA translesion synthesis, Adv. Exp. Med. Biol., 1241, 35-45, https://doi.org/10.1007/978-3-030-41283-8-3.
Maga, G., Crespan, E., Markkanen, E., Imhof, R., Furrer, A., Villani, G., Hübscher, U., and Van Loon, B. (2013) DNA polymerase δ-interacting protein 2 is a processivity factor for DNA polymerase λ during 8-oxo-7,8-dihydroguanine bypass, Proc. Natl. Acad. Sci. USA, 110, 18850-18855, https://doi.org/10.1073/pnas.1308760110.
Tissier, A., Janel-bintz, R., Coulon, S., Klaile, E., Kannouche, P., Fuchs, R. P. P., and Cordonnier, A. M. (2010) Crosstalk between replicative and translesional DNA polymerases: PDIP38 interacts directly with Pol eta, DNA Repair (Amst.), 9, 922-928, https://doi.org/10.1016/j.dnarep.2010.04.010.
Guilliam, T. A., Bailey, L. J., Brissett, N. C., and Doherty, A. J. (2016) PolDIP2 interacts with human PrimPol and enhances its DNA polymerase activities, Nucleic Acids Res., 44, 3317-3329, https://doi.org/10.1093/nar/gkw175.
Kazutoshi, K., Stojkovič, G., Velázquez-Ruiz, C., Martínez-Jiménez, M. I., Doimo, M., Laurent, T., Berner, A., Pérez-Rivera, A. E., Jenninger, L., Blanco, L., and Wanrooij, S. (2021) A unique arginine cluster in PolDIP2 enhances nucleotide binding and DNA synthesis by PrimPol, Nucleic Acids Res., 49, 2179-2191, https://doi.org/10.1093/nar/gkab049.
Tsuda, M., Ogawa, S., Ooka, M., Kobayashi, K., Hirota, K., Wakasugi, M., Matsunaga, T., Sakuma, T., Yamamoto, T., Chikuma, S., Sasanuma, H., Debatisse, M., Doherty, A. J., Fuchs, R. P., and Takeda, S. (2019) PDIP38/PolDIP2 controls the DNA damage tolerance pathways by increasing the relative usage of translesion DNA synthesis over template switching, PLoS One, 14, e0213383, https://doi.org/10.1371/journal.pone.0213383.
Taylor, M. R. G., and Yeeles, J. T. P. (2018) The initial response of a eukaryotic replisome to DNA damage, Mol. Cell, 70, 1067-1080, https://doi.org/10.1016/j.molcel.2018.04.022.
Lyu, K., Kumagai, A., and Dunphy, W. G. (2019) RPA-coated single-stranded DNA promotes the ETAA1-dependent activation of ATR, Cell Cycle, 18, 898-913, https://doi.org/10.1080/15384101.2019.1598728.
Van, C., Yan, S., Michael, W. M., Waga, S., and Cimprich, K. A. (2010) Continued primer synthesis at stalled replication forks contributes to checkpoint activation, J. Cell Biol., 189, 233-246, https://doi.org/10.1083/jcb.200909105.
Vallerga, M. B., Mansilla, S. F., Federico, M. B., Bertolin, A. P., and Gottifredi, V. (2015) Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation, Proc. Natl. Acad. Sci. USA, 112, 6624-6633, https://doi.org/10.1073/pnas.1508543112.
Bai, G., Kermi, C., Stoy, H., Schiltz, C. J., Bacal, J., Zaino, A. M., Hadden, M. K., Eichman, B. F., Lopes, M., and Cimprich, K. A. (2020) HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis, Mol. Cell, 78, 1237-1251.e7, https://doi.org/10.1016/j.molcel.2020.04.031.
Kang, Z., Fu, P., Alcivar, A. L., Fu, H., Redon, C., Foo, T. K., Zuo, Y., Ye, C., Baxley, R., Madireddy, A., Buisson, R., Bielinsky, A., Zou, L., Shen, Z., Aladjem, M. I., and Xia, B. (2021) BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage, Nat. Commun., 12, 5966, https://doi.org/10.1038/s41467-021-26227-6.
Couch, F. B., Bansbach, C. E., Driscoll, R., Luzwick, J. W., Glick, G. G., Bétous, R., Carroll, C. M., Jung, S. Y., Qin, J., Cimprich, K. A., and Cortez, D. (2013) ATR phosphorylates SMARCAL1 to prevent replication fork collapse, Genes Dev., 27, 1610-1623, https://doi.org/10.1101/gad.214080.113.
Berti, M., Chaudhuri, A. R., Thangavel, S., Gomathinayagam, S., Kenig, S., Vujanovic, M., Odreman, F., Glatter, T., Graziano, S., Mendoza-Maldonado, R., Marino, F., Lucic, B., Biasin, V., Gstaiger, M., Aebersold, R., Sidorova, J. M., Monnat, R. J. Jr., Lopes, M., and Vindigni, A. (2013) Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition, Nat. Struct. Mol. Biol., 20, 347-354, https://doi.org/10.1038/nsmb.2501.
Giansanti, C., Manzini, V., Dickmanns, A., Dickmanns, A., Palumbieri, M. D., Sanchi, A., Kienle, S. M., Rieth, S., Scheffner, M., Lopes, M., and Dobbelstein, M. (2022) MDM2 binds and ubiquitinates PARP1 to enhance DNA replication fork progression, Cell Rep., 39, 110879, https://doi.org/10.1016/j.celrep.2022.110879.
Genois, M. M., Gagné, J. P., Yasuhara, T., Jackson, J., Saxena, S., Langelier, M. F., Ahel, I., Bedford, M. T., Pascal, J. M., Vindigni, A., Poirier, G. G., and Zou, L. (2021) CARM1 regulates replication fork speed and stress response by stimulating PARP1, Mol. Cell, 81, 784-800.e8, https://doi.org/10.1016/j.molcel.2020.12.010.
Leuzzi, G., Marabitti, V., Pichierri, P., and Franchitto, A. (2016) WRNIP 1 protects stalled forks from degradation and promotes fork restart after replication stress, EMBO J., 35, 1437-1451, https://doi.org/10.15252/embj.201593265.
Taglialatela, A., Leuzzi, G., Sannino, V., Cuella-Martin, R., Huang, J. W., Wu-Baer, F., Baer, R., Costanzo, V., and Ciccia, A. (2021) REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps, Mol. Cell, 81, 4008-4025.e7, https://doi.org/10.1016/j.molcel.2021.08.016.
Li, J., and Zhang, Q. (2015) Primpol mutation: functional study does not always reveal the truth, Investig. Ophthalmol. Vis. Sci., 56, 1181-1182, https://doi.org/10.1167/iovs.14-16072.
Yuan, H., Wang, Q., Li, Y., Cheng, S., Liu, J., and Liu, Y. (2020) Concurrent pathogenic variants in SLC6A1/NOTCH1/PRIMPOL genes in a Chinese patient with myoclonic-atonic epilepsy, mild aortic valve stenosis and high myopia, BMC Med. Genet., 21, 93, https://doi.org/10.1186/s12881-020-01035-9.
Pilzecker, B., Buoninfante, O. A., Pritchard, C., Blomberg, O. S., Huijbers, I. J., Van Den Berk, P. C. M., and Jacobs, H. (2016) PrimPol prevents APOBEC/AID family mediated DNA mutagenesis, Nucleic Acids Res., 44, 4734-4744, https://doi.org/10.1093/nar/gkw123.
Díaz-Talavera, A., Montero-Conde, C., Leandro-García, L. J., and Robledo, M. (2022) PrimPol: a breakthrough among DNA replication enzymes and a potential new target for cancer therapy, Biomolecules, 12, 248, https://doi.org/10.3390/biom12020248.
Acknowledgments
The authors are grateful to Yu. I. Pavlov and A. G. Baranovskiy for valuable discussions and advice.
Funding
This work was supported by the Russian Science Foundation (grant no. 22-24-20150 to E.O.B).
Author information
Authors and Affiliations
Contributions
E.O.B. figure preparation, writing the draft of the paper; A.V.M. draft correction.
Corresponding author
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain studies involving human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Boldinova, E.O., Makarova, A.V. Regulation of Human DNA Primase-Polymerase PrimPol. Biochemistry Moscow 88, 1139–1155 (2023). https://doi.org/10.1134/S0006297923080084
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297923080084