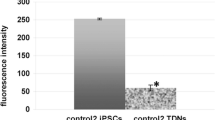
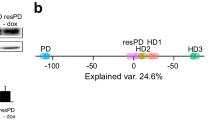
Abstract
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases in the world. Despite numerous studies, the causes of this pathology remain completely unknown. This is, among other things, due to the difficulty of obtaining biological material for analysis. Neural cell cultures derived from the induced pluripotent stem cells (IPSCs) provide a great potential for studying molecular events underlying the pathogenesis of PD. This paper presents the results of bioinformatic analysis of the data obtained using RNA-seq technology in the study of neural precursors (NP) derived from IPSCs of the healthy donors and patients with PD carrying various mutations that are commonly associated with familial PD. This analysis showed that the level of transcription of multiple genes actively expressed in the nervous system at the embryonic stage of development was significantly increased in the NP cells obtained from the patients with PD, unlike in the case of healthy donors. Bioinformatic data have been, in general, confirmed using real-time PCR. The obtained data suggest that one of the causes of PD may be the shift of the gene expression pattern in neuronal cells towards embryonic gene expression pattern (termed dematuration).



Similar content being viewed by others
Abbreviations
- DA neurons:
-
dopaminergic neurons
- DEGs:
-
differentially expressed genes
- FDR:
-
false discovery rate
- HD:
-
healthy donor
- IPSC:
-
induced pluripotent stem cells
- NP:
-
neural precursors
- PD:
-
Parkinson’s disease
References
De Lau, L. M., and Breteler, M. M. B. (2006) Epidemiology of Parkinson’s disease, Lancet Neurol., 5, 525-535, https://doi.org/10.1016/S1474-4422(06)70471-9.
Okano, H., and Yamanaka, S. (2014) iPS cell technologies: significance and applications to CNS regeneration and disease, Mol. Brain, 7, 22, https://doi.org/10.1186/1756-6606-7-22.
Day, J. O., and Mullin, S. (2021) The genetics of Parkinson’s disease and implications for clinical practice, Genes (Basel), 12, 1006, https://doi.org/10.3390/genes12071006.
Rathore, A. S., Birla, H., Singh, S. S., Zahra, W., Dilnashin, H., Singh, R., Keshri, P. K., and Singh, S. P. (2021) Epigenetic modulation in Parkinson’s disease and potential treatment therapies, Neurochem. Res., 46, 1618-1626, https://doi.org/10.1007/s11064-021-03334-w.
Rouaud, T., Corbillé, A.-G., Leclair-Visonneau, L., de Lataillade, A. G., Lionnet, A., Preterre, C., Damier, P., and Derkinderen, P. (2021) Pathophysiology of Parkinson’s disease: mitochondria, alpha-synuclein and much more, Rev. Neurol. (Paris), 177, 260-271, https://doi.org/10.1016/j.neurol.2020.07.016.
Novosadova, E. V., and Grivennikov, I. A. (2014) Induced pluripotent stem cells: from derivation to application in biochemical and biomedical research, Biochemistry (Moscow), 79, 1425-1441, https://doi.org/10.1134/S000629791413001X.
Novosadova, E. D., Nekrasov, E. D., Chestkov, I. V., Surdina, A. V., Vasina, E. M., Bogomazova, A. N., Manuilova, E. S., Arsen’eva, E. L., Simonova, V. V., Konovalova, E. V., Fedotova, E. Yu., Abramicheva, N. Yu., Khaspekov, L. G., Grivennikov, I. A., Tarantul, V. Z., Kiselev, S. L., and Illarioshkin, S. N. (2016) A platform for studying molecular and cellular mechanisms of Parkinson’s disease based on human induced pluripotent stem cells [in Russian], Sovrem. Tehnol. Med., 8, 157-166, https://doi.org/10.17691/stm2016.8.4.20.
Fedoseeva, V. B., Novosadova, E. V., Novosadova, V. V., Nenasheva, V. V., Grivennikov, I. A., and Tarantul, V. Z. (2022) The level of transcription of HOX genes increased in neural precursors derived from iPSC from patients with Parkinson’s disease, in Smart and Innovative Farming for Sustainable Agriculture and Food Systems, Springer Nature.
Novosadova, E., Anufrieva, K., Kazantseva, E., Arsenyeva, E., Fedoseyeva, V., Stepanenko, E., Poberezhniy, D., Illarioshkin, S., Novosadova, L., Gerasimova, T., Nenasheva, V., Grivennikov, I., Lagarkova, M., and Tarantul, V. (2022) Transcriptome datasets of neural progenitors and neurons differentiated from induced pluripotent stem cells of healthy donors and Parkinson’s disease patients with mutations in the PARK2 gene, Data Brief, 41, 107958, https://doi.org/10.1016/j.dib.2022.107958.
Avazzadeh, S., Baena, J. M., Keighron, C., Feller-Sanchez, Y., and Quinlan, L. R. (2021) Modelling Parkinson’s disease: iPSCs towards better understanding of human pathology, Brain Sci., 11, 373, https://doi.org/10.3390/brainsci11030373.
Novosadova, E. V., Nenasheva, V. V., Makarova, I. V., Dolotov, O. V., Inozemtseva, L. S., Arsenyeva, E. L., Chernyshenko, S. V., Sultanov, R. I., Illarioshkin, S. N., Grivennikov, I. A., and Tarantul, V. Z. (2020) Parkinson’s disease-associated changes in the expression of neurotrophic factors and their receptors upon neuronal differentiation of human induced pluripotent stem cells, J. Mol. Neurosci., 70, 514-521, https://doi.org/10.1007/s12031-019-01450-5.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010) EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 26, 139-140, https://doi.org/10.1093/bioinformatics/btp616.
Livak, K., and Schmittgen, T. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method, Methods, 25, 402-408, https://doi.org/10.1006/meth.2001.1262.
Kulakovskiy, I. V., Vorontsov, I. E., Yevshin, I. S., Sharipov, R. N., Fedorova, A. D., Rumynskiy, E. I., Medvedeva, Y. A., Magana-Mora, A., Bajic, V. B., Papatsenko, D. A., Kolpakov, F. A., and Makeev, V. J. (2018) HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis, Nucleic Acids Res., 46(D1), D252-D259, https://doi.org/10.1093/nar/gkx1106.
Wang, W. D., Melville, D. B., Montero-Balaguer, M., Hatzopoulos, A. K., and Knapik, E. W. (2011) Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population, Dev. Biol., 360, 173-185, https://doi.org/10.1016/j.ydbio.2011.09.0194.
Hulme, A. J., Maksour, S., St-Clair Glover, M., Miellet, S., and Dottori, M. (2022) Making neurons, made easy: the use of neurogenin-2 in neuronal differentiation, Stem Cell Rep., 17, 14-34, https://doi.org/10.1016/j.stemcr.2021.11.015.
Ang, S.-L. (2009) Foxa1 and Foxa2 transcription factors regulate differentiation of midbrain dopaminergic neurons, Adv. Exp. Med. Biol., 651, 58-65, https://doi.org/10.1007/978-1-4419-0322-8_5.
Blesa, J., and Przedborski, S. (2014) Parkinson’s disease: animal models and dopaminergic cell vulnerability, Front. Neuroanat., 8, 155-167, https://doi.org/10.3389/fnana.2014.00155.
Takahashi, K., and Yamanaka, S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell, 126, 663-676, https://doi.org/10.1016/j.cell.2006.07.024.
Marchionini, M., Lehrmann, E., Chu, Y., He, B., Sorwell, C. E., Beckerc, K. G., Freed, W. J., Kordower, J. H., and Collier, T. J. (2007) Role of heparin binding growth factors in nigrostriatal dopamine system development and Parkinson’s disease, Brain Res., 1147, 77-88, https://doi.org/10.1016/j.brainres.2007.02.028.
Zagare, A., Barmpa, K., Smajic, S., Smits, L. M., Grzyb, K., Grünewald, A., Skupin, A., Nickels, S. L., and Schwamborn, J. C. (2022) Midbrain organoids mimic early embryonic neurodevelopment and recapitulate LRRK2-p.Gly2019Ser-associated gene expression, Am. J. Hum. Genet., 109, 311-327, https://doi.org/10.1016/j.ajhg.2021.12.009.
Wang, Y., and Wang, Z. (2020) An integrated network analysis of mRNA and gene expression profiles in Parkinson’s disease, Med. Sci. Monit., 26, 920846, https://doi.org/10.12659/msm.920846.
Bansod, S., Kageyama, R., and Ohtsuka, T. (2017) HES5 regulates the transition timing of neurogenesis and gliogenesis in mammalian neocortical development, Development, 144, 3156-3167, https://doi.org/10.1242/dev.147256.
Chapman, G., Sparrow, D. B., Kremmer, E., and Dunwoodie, S. L. (2011) Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis, Hum. Mol. Genet., 20, 905-916, https://doi.org/10.1093/hmg/ddq529.
Augustyn, A., Borromeo, M., Wang, T., Fujimoto, J., Shao, C., Dospoy, P. D., Lee, V., Tan, C., Sullivan, J. P., Larsen, J. E., Girard, L., Behrens, C., Wistuba, I. I., Xie, Y., Cobb, M. H., Gazdar, A. F., Johnson, J. E., and Minna, J. D. (2014) ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers, Proc. Natl. Acad. Sci. USA, 111, 14788-14793, https://doi.org/10.1073/pnas.1410419111.
Henke, R. M., Meredith, D. M., Borromeo, M. D., Savage, T. K., and Johnson, J. E. (2009) Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube, Dev. Biol., 328, 529-540, https://doi.org/10.1016/j.ydbio.2009.01.007.
Kageyama, R., Shimojo, H., and Ohtsuka, T. (2019) Dynamic control of neural stem cells by bHLH factors, Neurosci. Res., 138, 12-18, https://doi.org/10.1016/j.neures.2018.09.005.
Ide, M., Yamada, K., Toyota, T., Iwayama, Y., Ishitsuka, Y., Minabe, Y., Nakamura, K., Hattori, N., Asada, T., Mizuno, Y., Mori, N., and Yoshikawa, T. (2005) Genetic association analyses of PHOX2B and ASCL1 in neuropsychiatric disorders: evidence for association of ASCL1 with Parkinson’s disease, Hum. Genet., 117, 520-527, https://doi.org/10.1007/s00439-005-1342-8.
Oliveira, M. A. P., Balling, R., Smidt, M. P., and Fleming, R. M. T. (2017) Embryonic development of selectively vulnerable neurons in Parkinson’s disease, NPJ Parkinson Dis., 3, 21, https://doi.org/10.1038/s41531-017-0022-4.
Caiazzo, M., Dell’Anno, M. T., Dvoretskova, E., Lazarevic, D., Taverna, S., Leo, D., Sotnikova, T. D., Menegon, A., Roncaglia, P., Colciago, G., Russo, G., Carninci, P., Pezzoli, G., Gainetdinov, R. R., Gustincich, S., Dityatev, A., and Broccoli, V. (2011) Direct generation of functional dopaminergic neurons from mouse and human fibroblasts, Nature, 476, 224-227, https://doi.org/10.1038/nature10284.
Lu, C., Shi, X., Allen, A., Baez-Nieto, D., Nikish, A., Sanjana, N. E., and Pan, J. Q. (2019) Overexpression of NEUROG2 and NEUROG1 in human embryonic stem cells produces a network of excitatory and inhibitory neurons, FASEB J., 33, 5287-5299, https://doi.org/10.1096/fj.201801110RR.
Han, S., Dennis, D. F., Balakrishnan, A., Dixit, R., Britz, O., Zinyk, D., Touahri, Y., Olender, T., Brand, M., Guillemot, F., Kurrasch, D., and Schuurmans, C. (2018) A non-canonical role for the proneural gene Neurog1 as a negative regulator of neocortical neurogenesis, Development, 145, dev157719, https://doi.org/10.1242/dev.157719.
Kele, J., Simplicio, N., Ferri, A. L. M., Mira, H., Guillemot, F., Arenas, E., and Ang, S.-L. (2006) Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons, Development, 133, 495-505, https://doi.org/10.1242/dev.02223.
Dasen, J. S., Tice, B. C., Brenner-Morton, S., and Jessell, T. M. (2005) A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity, Cell, 123, 477-491, https://doi.org/10.1016/j.cell.2005.09.009.
Schmid, T., Krüger, M., and Braun, T. (2007) NSCL-1 and -2 control the formation of precerebellar nuclei by orchestrating the migration of neuronal precursor cells, Neurochemistry, 102, 2061-2072, https://doi.org/10.1111/j.1471-4159.2007.04694.x.
Kratochwil, C. F., Maheshwari, U., and Rijli, F. M. (2017) The long journey of pontine nuclei neurons: from rhombic lip to cortico-ponto-cerebellar circuitry, Front. Neurol. Circuits, 11, 33, https://doi.org/10.3389/fncir.2017.00033.
Krüger, M., Ruschke, K., and Braun, T. (2004) NSCL-1 and NSCL-2 synergistically determine the fate of GnRH-1 neurons and control necdin gene expression, EMBO J., 23, 4353-4364, https://doi.org/10.1038/sj.emboj.7600431.
Byun, J. S., Oh, M., Lee, S., Gil, J.-E., Mo, Y., Ku, B., Kim, W. K., Oh, K. J., Lee, E. W., Bae, K. H., Lee, S. C., and Han, B. S. (2020) The transcription factor PITX1 drives astrocyte differentiation by regulating the SOX9 gene, J. Biol. Chem., 295, 13677-13690, https://doi.org/10.1074/jbc.RA120.013352.
Agoston, Z., Li, N., Haslinger, A., Wizenmann, A., and Schulte, D. (2012) Genetic and physical interaction of Meis2, Pax3 and Pax7 during dorsal midbrain development, BMC Dev. Biol., 12, 10, https://doi.org/10.1186/1471-213X-12-10.
Mansouri, A. (1998) The role of Pax3 and Pax7 in development and cancer, Crit. Rev. Oncog., 9, 141-149, https://doi.org/10.1615/critrevoncog.v9.i2.40.
Agoston, Z., and Schulte, D. (2009) Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer, Development, 136, 3311-3322, https://doi.org/10.1242/dev.037770.
Xu, M., Li, Y., Du, J., Lin, H., Cao, S., Mao, Z., Wu, R., Liu, M., Liu, Y., and Yin, Q. (2018) PAX3 promotes cell migration and CXCR4 gene expression in neural crest cells, J. Mol. Neurosci., 64, 1-8, https://doi.org/10.1007/s12031-017-0995-9.
Green, Y. S., and Vetter, M. L. (2021) EBF factors drive expression of multiple classes of target genes governing neuronal development, Neural. Dev., 6, 19, https://doi.org/10.1186/1749-8104-6-19.
Chandrasekaran, S., and Bonchev, D. (2013) A network view on Parkinson’s disease, Comput. Struct. Biotechnol., 7, 201304004, https://doi.org/10.5936/csbj.201304004.
Cruz-Monteagudo, M., Borges, F., Paz-Y-Miño, C., Cordeiro, M. N. D. S., Rebelo, I., Perez-Castillo, Y., Helguera, A. M., Sánchez-Rodríguez, A., and Tejera, E. (2016) Efficient and biologically relevant consensus strategy for Parkinson’s disease gene prioritization, BMC Med. Genomics, 9, 12, https://doi.org/10.1186/s12920-016-0173-x.
Maden, M. (2007) Retinoic acid in the development, regeneration and maintenance of the nervous system, Nat. Rev. Neurosci., 8, 755-765, https://doi.org/10.1038/nrn2212.
Napoli, J. L. (2012) Physiological insights into all-trans-retinoic acid biosynthesis, Biochim. Biophys. Acta, 1821, 152-167, https://doi.org/10.1016/j.bbalip.2011.05.004.
Yamamoto, M., Zhang, J., Smith, D., Hayakawa, Y., and McCaffery, P. (2003) A critical period for retinoic acid teratogenesis and loss of neurophilic migration of pontine nuclei neurons, Mech. Dev., 120, 701-709, https://doi.org/10.1016/s0925-4773(03)00047-9.
Yamamoto, M., Fujinuma, M., Hirano, S., Hayakawa, Y., Clagett-Dame, M., Zhang, J., and McCaffery, P. (2005) Retinoic acid influences the development of the inferior olivary nucleus in the rodent, Dev. Biol., 280, 421-433, https://doi.org/10.1016/j.ydbio.2005.02.007.
Bouillet, P., Chazaud, C., Oulad-Abdelghani, M., Dollé, P., and Chambon, P. (1995) Sequence and expression pattern of the Stra7 (Gbx-2) homeobox-containing gene induced by retinoic acid in P19 embryonal carcinoma cells, Dev. Dyn., 204, 372-382, https://doi.org/10.1002/aja.1002040404.
Wassarman, K. M., Lewandoski, M., Campbell, K., Joyner, A. L., Rubenstein, J. L., Martinez, S., and Martin, G. R. (1997) Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function, Development, 124, 2923-2934, https://doi.org/10.1242/dev.124.15.2923.
Millet, S., Campbell, K., Epstein, D. J., Losos, K., Harris, E., and Joyner, A. (1999) A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer, Nature, 401, 161-164, https://doi.org/10.1038/43664.
Mesman, S., and Smidt, M. P. (2020) Acquisition of the midbrain dopaminergic neuronal identity, Int. J. Mol. Sci., 21, 4638, https://doi.org/10.3390/ijms21134638.
Prakash, N., Brodski, C., Naserke, T., Puelles, E., Gogoi, R., Hall, A., Panhuysen, M., Echevarria, D., Sussel, L., Weisenhorn, D. M., Martinez, S., Arenas, E., Simeone, A., and Wurst, W. (2006) A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo, Development, 133, 89-98, https://doi.org/10.1242/dev.02181.
Yang, J., Brown, A., Ellisor, D., Paul, E., Hagan, N., and Zervas, M. (2013) Dynamic temporal requirement of Wnt1 in midbrain dopamine neuron development, Development, 140, 1342-1352, https://doi.org/10.1242/dev.080630.
Arenas, E. (2014) Wnt signaling in midbrain dopaminergic neuron development and regenerative medicine for Parkinson’s disease, J. Mol. Cell. Biol., 6, 42-53, https://doi.org/10.1093/jmcb/mju001.
Akane, H., Saito, F., Shiraki, A., Imatanaka, N., Akahori, Y., Itahashi, M., Wang, L., and Shibutani, M. J. (2014) Gene expression profile of brain regions reflecting aberrations in nervous system development targeting the process of neurite extension of rat offspring exposed developmentally to glycidol, Appl. Toxicol., 34, 1389-1399, https://doi.org/10.1002/jat.2971.
Rawal, N., Corti, O., Sacchetti, P., Ardilla-Osorio, H., Sehat, B., Brice, A., and Arenas, E. (2009) Parkin protects dopaminergic neurons from excessive Wnt/beta-catenin signaling, Biochem. Biophys. Res. Commun., 388, 473-478, https://doi.org/10.1016/j.bbrc.2009.07.014.
Sancho, R. M., Law, B. M. H., and Harvey, K. (2009) Mutations in the LRRK2 Roc-COR tandem domain link Parkinson’s disease to Wnt signaling pathways, Hum. Mol. Genet., 18, 3955-3968, https://doi.org/10.1093/hmg/ddp337.
Berwick, D. C., and Harvey, K. (2012) LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6, Hum. Mol. Genet., 21, 4966-4979, https://doi.org/10.1093/hmg/dds342.
Rothstein, M., and Simoes-Costa, M. (2020) Heterodimerization of TFAP2 pioneer factors drives epigenomic remodeling during neural crest specification, Genome Res., 30, 35-48, https://doi.org/10.1101/gr.249680.119.
Kousa, Y. A., Zhu, H., Fakhouri, W. D., Lei, Y., Kinoshita, A., Roushangar, R. R., Patel, N. K., Agopian, A. J., Yang, W., Leslie, E. J., Busch, T. D., Mansour, T. A., Li, X., Smith, A. L., Li, E. B., Sharma, D. B., Williams, T. J., Chai, Y., Amendt, B. A., Liao, E. C., Mitchell, L. E., Bassuk, A. G., Gregory, S., Ashley-Koch, A., Shaw, G. M., Finnell, R. H., and Schutte, B. C. (2019) TFAP2A–IRF6–GRHL genetic pathway is conserved in neurulation, Hum. Mol. Genet., 28, 1726-1737, https://doi.org/10.1093/hmg/ddz010.
Ahn, J.-I., Lee, K.-H., Shin, D.-M., Shim, J.-W., Lee, J.-S., Chang, S.Y., Lee, Y.-S., Brownstein, M. J., Lee, S.-H., and Lee, Y.-S. (2004) Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons, Dev. Biol., 265, 491-501, https://doi.org/10.1016/j.ydbio.2003.09.041.
Sim, H., Lee, J.-E., Yoo, H. M., Cho, S., Lee, H., Baek, A., Kim, J., Seo, H., Kweon, M. N., Kim, H. G., Jeon, Y. J., Son, M. Y., and Kim, J. (2020) Iroquois homeobox protein 2 identified as a potential biomarker for Parkinson’s disease, Int. J. mol. Sci., 21, 3455, https://doi.org/10.3390/ijms21103455.
Hagihara, H., Ohira, K., and Miyakawa, T. (2019) Transcriptomic evidence for immaturity induced by antidepressant fluoxetine in the hippocampus and prefrontal cortex, Neuropsychopharmacol. Rep., 39, 78-89, https://doi.org/10.1002/npr2.12048.
Hagihara, H., Murano, T., Ohira, K., Miwa, M., Nakamura, K., and Miyakawa, T. (2019) Expression of progenitor cell/immature neuron markers does not present definitive evidence for adult neurogenesis, Mol. Brain, 12, 108, https://doi.org/10.1186/s13041-019-0522-8.
Lehrer, S., and Rheinstein, P. H. (2021) Alzheimer’s disease and Parkinson’s disease may result from reactivation of embryologic pathways silenced at birth, Discov. Med., 31, 89-94.
Caldwell, A. B., Liu, Q., Schroth, G. P., Galasko, D. R., Yuan, S. H., Wagner, S. T., and Subramaniam, S. (2020) Dedifferentiation and neuronal repression define familial Alzheimer’s disease, Sci. Adv., 6, eaba5933, https://doi.org/10.1126/sciadv.aba5933.
Weykopf, B., Haupt, S., Jungverdorben, J., Flitsch, L. J., Hebisch, M., Liu, G.-H., Suzuki, K., Belmonte, J. C. I., Peitz, M., Blaess, S., Till, A., and Brüstle, O. (2019) Induced pluripotent stem cell-based modeling of mutant LRRK2-associated Parkinson’s disease, Eur. J. Neurosci., 49, 561-589, https://doi.org/10.1111/ejn.14345.
Acknowledgments
The authors express their gratitude to academician S. N. Illarioshkin (Research Center of Neurology) for providing biopsy sample from the patients with PD, to professor M. A. Lagarkova (Federal Research and Clinical Center of Physical Chemical Medicine of the Federal Medical and Biological Agency of the Russian Federation, Moscow, Russia) for providing IPSC cell lines. Experiments were conducted with the equipment of the Center of Collective Use of scientific equipment (Center “Cellular and Genetic Technologies” of Institute of Molecular Genetics of National Research Centre “Kurchatov Institute”).
Funding
The study was financially supported by the Ministry of Science and Higher Education of the Russian Federation (agreement no. 075-15-2021-1357) (generation of cell lines) and by the Russian Science Foundation (project no. 21-15-00103) (bioinformatic analysis).
Author information
Authors and Affiliations
Contributions
All authors provided contribution to the development of concept and design of the study. E. V. Novosadova and V. V. Nenasheva prepared material for examination; E. V. Novosadova, L. V. Novosadova, V. V. Nenasheva, I. A. Grivennikov, and V. B. Fedoseyeva collected and analyzed the data. First draft of the paper was prepared by V. B. Fedoseyeva and V. Z. Tarantul. All authors approved final version of the manuscript.
Corresponding author
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors. Description of cell lines produced from human cells obtained with their consent is presented in our previous papers, where these cell lines were first introduced.
Rights and permissions
About this article
Cite this article
Fedoseyeva, V.B., Novosadova, E.V., Nenasheva, V.V. et al. Activation of Embryonic Gene Transcription in Neural Precursor Cells Derived from the Induced Pluripotent Stem Cells of the Patients with Parkinson’s Disease. Biochemistry Moscow 88, 515–525 (2023). https://doi.org/10.1134/S0006297923040077
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297923040077