Abstract
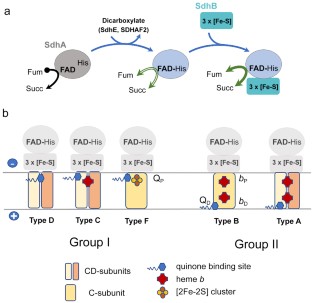
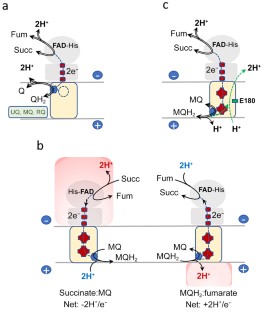
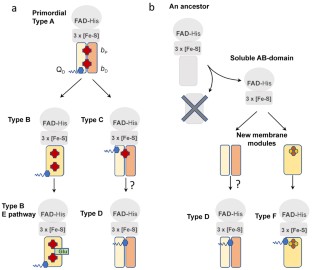
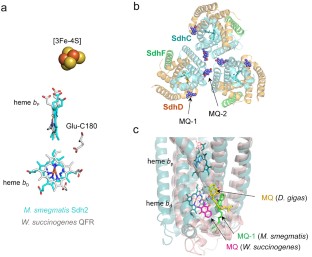
The Complex II family encompasses membrane bound succinate:quinones reductases and quinol:fumarate reductases that catalyze interconversion of succinate and fumarate coupled with reduction and oxidation of quinone. These enzymes are found in all biological genres and share a modular structure where a highly conserved soluble domain is bound to a membrane-spanning domain that is represented by distinct variations. The current classification of the complex II family members is based on the number of subunits and co-factors in the membrane anchor (types A-F). This classification also provides insights into possible evolutionary paths and suggests that some of the complex II enzymes (types A-C) co-evolved as the whole assembly. Origin of complex II types D and F may have arisen from independent events of de novo association of the conserved soluble domain with a new anchor. Here we analyze a recent structure of Mycobacterium smegmatis Sdh2, a complex II enzyme with two transmembrane subunits and two heme b molecules. This analysis supports an earlier hypothesis suggesting that mitochondrial complex II (type C) with a single heme b may have evolved as an assembled unit from an ancestor similar to M. smegmatis Sdh2.
Similar content being viewed by others
Abbreviations
- CII:
-
complex II
- cryo-EM:
-
cryo-electron microscopy
- heme b D :
-
distal heme b
- heme b P :
-
proximal heme b
- QFR:
-
quinol:fumarate oxidoreductase
- QD :
-
distal quinone binding site
- QP :
-
proximal quinone binding site
- SQR:
-
succinate:quinone reductase
References
Cecchini, G. (2003) Function and structure of complex II of the respiratory chain, Annu. Rev. Biochem., 72, 77-109, https://doi.org/10.1146/annurev.biochem.72.121801.161700.
Hagerhall, C. (1997) Succinate: quinone oxidoreductases. Variations on a conserved theme, Biochim. Biophys. Acta, 1320, 107-141, https://doi.org/10.1016/s0005-2728(97)00019-4.
Sharma, P., Maklashina, E., Cecchini, G., and Iverson, T. M. (2019) Maturation of the respiratory complex II flavoprotein, Curr. Opin. Struct. Biol., 59, 38-46, https://doi.org/10.1016/j.sbi.2019.01.027.
Sharma, P., Maklashina, E., Cecchini, G., and Iverson, T. M. (2018) Crystal structure of an assembly intermediate of respiratory Complex II, Nat. Commun., 9, 274, https://doi.org/10.1038/s41467-017-02713-8.
Kounosu, A. (2014) Analysis of covalent flavinylation using thermostable succinate dehydrogenase from Thermus thermophilus and Sulfolobus tokodaii lacking SdhE homologs, FEBS Lett., 588, 1058-1063, https://doi.org/10.1016/j.febslet.2014.02.022.
Lill, R., and Freibert, S. A. (2020) Mechanisms of mitochondrial iron-sulfur protein biogenesis, Annu. Rev. Biochem., 89, 471-499, https://doi.org/10.1146/annurev-biochem-013118-111540.
Bai, Y., Chen, T., Happe, T., Lu, Y., and Sawyer, A. (2018) Iron-sulphur cluster biogenesis via the SUF pathway, Metallomics, 10, 1038-1052, https://doi.org/10.1039/c8mt00150b.
Heuts, D. P., Scrutton, N. S., McIntire, W. S., and Fraaije, M. W. (2009) What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors, FEBS J., 276, 3405-3427, https://doi.org/10.1111/j.1742-4658.2009.07053.x.
Hao, H. X., Khalimonchuk, O., Schraders, M., Dephoure, N., Bayley, J. P., et al. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma, Science, 325, 1139-1142, https://doi.org/10.1126/science.1175689.
McNeil, M. B., Clulow, J. S., Wilf, N. M., Salmond, G. P., and Fineran, P. C. (2012) SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria, J. Biol. Chem., 287, 18418-18428, https://doi.org/10.1074/jbc.M111.293803.
Van Vranken, J. G., Na, U., Winge, D. R., and Rutter, J. (2015) Protein-mediated assembly of succinate dehydrogenase and its cofactors, Crit. Rev. Biochem. Mol. Biol., 50, 168-180, https://doi.org/10.3109/10409238.2014.990556.
Moosavi, B., Berry, E. A., Zhu, X. L., Yang, W. C., and Yang, G. F. (2019) The assembly of succinate dehydrogenase: a key enzyme in bioenergetics, Cell. Mol. Life Sci., 76, 4023-4042, https://doi.org/10.1007/s00018-019-03200-7.
Tedeschi, G., Negri, A., Mortarino, M., Ceciliani, F., Simonic, T., et al. (1996) L-aspartate oxidase from Escherichia coli. II. Interaction with C4 dicarboxylic acids and identification of a novel L-aspartate: fumarate oxidoreductase activity, Eur. J. Biochem., 239, 427-433, https://doi.org/10.1111/j.1432-1033.1996.0427u.x.
Taylor, P., Pealing, S. L., Reid, G. A., Chapman, S. K., and Walkinshaw, M. D. (1999) Structural and mechanistic mapping of a unique fumarate reductase, Nat. Struct. Biol., 6, 1108-1112, https://doi.org/10.1038/70045.
Maklashina, E., Rajagukguk, S., Iverson, T. M., and Cecchini, G. (2018) The unassembled flavoprotein subunits of human and bacterial complex II have impaired catalytic activity and generate only minor amounts of ROS, J. Biol. Chem., 293, 7754-7765, https://doi.org/10.1074/jbc.RA118.001977.
Maklashina, E., Iverson, T. M., Sher, Y., Kotlyar, V., Andrell, J., et al. (2006) Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain, J. Biol. Chem., 281, 11357-11365, https://doi.org/10.1074/jbc.M512544200.
Hägerhäll, C., and Hederstedt, L. (1996) A structural model for the membrane-integral domain of succinate: quinone oxidoreductases, FEBS Lett., 389, 25-31, https://doi.org/10.1016/0014-5793(96)00529-7.
Lancaster, C. R. (2013) The di-heme family of respiratory complex II enzymes, Biochim. Biophys. Acta, 1827, 679-687, https://doi.org/10.1016/j.bbabio.2013.02.012.
Iverson, T. M., Luna-Chavez, C., Cecchini, G., and Rees, D. C. (1999) Structure of the Escherichia coli fumarate reductase respiratory complex, Science, 284, 1961-1966, https://doi.org/10.1126/science.284.5422.1961.
Yankovskaya, V., Horsefield, R., Tornroth, S., Luna-Chavez, C., Miyoshi, H., et al. (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation, Science, 299, 700-704, https://doi.org/10.1126/science.1079605.
Huang, L. S., Shen, J. T., Wang, A. C., and Berry, E. A. (2006) Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the “oxaloacetate-inhibited” state, Biochim. Biophys. Acta, 1757, 1073-1083, https://doi.org/10.1016/j.bbabio.2006.06.015.
Sun, F., Huo, X., Zhai, Y., Wang, A., Xu, J., et al. (2005) Crystal structure of mitochondrial respiratory membrane protein complex II, Cell, 121, 1043-1057, https://doi.org/10.1016/j.cell.2005.05.025.
Inaoka, D. K., Shiba, T., Sato, D., Balogun, E. O., Sasaki, T., et al. (2015) Structural insights into the molecular design of flutolanil derivatives targeted for fumarate respiration of parasite mitochondria, Int. J. Mol. Sci., 16, 15287-15308, https://doi.org/10.3390/ijms160715287.
Hards, K., Rodriguez, S. M., Cairns, C., and Cook, G. M. (2019) Alternate quinone coupling in a new class of succinate dehydrogenase may potentiate mycobacterial respiratory control, FEBS Lett., 593, 475-486, https://doi.org/10.1002/1873-3468.13330.
Zhou, X., Gao, Y., Wang, W., Yang, X., Yang, X., et al. (2021) Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster, Proc. Natl. Acad. Sci. USA, 118, e2022308118, https://doi.org/10.1073/pnas.2022308118.
Lancaster, C. R., Kroger, A., Auer, M., and Michel, H. (1999) Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution, Nature, 402, 377-385, https://doi.org/10.1038/46483.
Guan, H. H., Hsieh, Y. C., Lin, P. J., Huang, Y. C., Yoshimura, M., et al. (2018) Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas, Sci. Rep., 8, 14935, https://doi.org/10.1038/s41598-018-33193-5.
Gong, H., Gao, Y., Zhou, X., Xiao, Y., Wang, W., et al. (2020) Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF, Nat. Commun., 11, 4245, https://doi.org/10.1038/s41467-020-18011-9.
Juhnke, H. D., Hiltscher, H., Nasiri, H. R., Schwalbe, H., and Lancaster, C. R. (2009) Production, characterization and determination of the real catalytic properties of the putative “succinate dehydrogenase” from Wolinella succinogenes, Mol. Microbiol., 71, 1088-1101, https://doi.org/10.1111/j.1365-2958.2008.06581.x.
Tran, Q. M., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2007) Escherichia coli succinate dehydrogenase variant lacking the heme b, Proc. Natl. Acad. Sci. USA, 104, 18007-18012, https://doi.org/10.1073/pnas.0707732104.
Maklashina, E., Hellwig, P., Rothery, R. A., Kotlyar, V., Sher, Y., et al. (2006) Differences in protonation of ubiquinone and menaquinone in fumarate reductase from Escherichia coli, J. Biol. Chem., 281, 26655-26664, https://doi.org/10.1074/jbc.M602938200.
Schirawski, J., and Unden, G. (1998) Menaquinone-dependent succinate dehydrogenase of bacteria catalyzes reversed electron transport driven by the proton potential, Eur. J. Biochem, 257, 210-215, https://doi.org/10.1046/j.1432-1327.1998.2570210.x.
Madej, M. G., Nasiri, H. R., Hilgendorff, N. S., Schwalbe, H., Unden, G., et al. (2006) Experimental evidence for proton motive force-dependent catalysis by the diheme-containing succinate:menaquinone oxidoreductase from the Gram-positive bacterium Bacillus licheniformis, Biochemistry, 45, 15049-15055, https://doi.org/10.1021/bi0618161.
Madej, M. G., Nasiri, H. R., Hilgendorff, N. S., Schwalbe, H., and Lancaster, C. R. (2006) Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex, EMBO J., 25, 4963-4970, https://doi.org/10.1038/sj.emboj.7601361.
Schafer, G., Anemuller, S., and Moll, R. (2002) Archaeal complex II: “classical” and “non-classical” succinate:quinone reductases with unusual features, Biochim. Biophys. Acta, 1553, 57-73, https://doi.org/10.1016/s0005-2728(01)00232-8.
Maklashina, E., Rajagukguk, S., McIntire, W. S., and Cecchini, G. (2010) Mutation of the heme axial ligand of Escherichia coli succinate-quinone reductase: implications for heme ligation in mitochondrial complex II from yeast, Biochim. Biophys. Acta, 1797, 747-754, https://doi.org/10.1016/j.bbabio.2010.01.019.
Lancaster, C. R., Gorss, R., Haas, A., Ritter, M., Mantele, W., et al. (2000) Essential role of Glu-C66 for menaquinol oxidation indicates transmembrane electrochemical potential generation by Wolinella succinogenes fumarate reductase, Proc. Natl. Acad. Sci. USA, 97, 13051-13056, https://doi.org/10.1073/pnas.220425797.
Maklashina, E., and Cecchini, G. (2010) The quinone-binding and catalytic site of complex II, Biochim. Biophys. Acta, 1797, 1877-1882, https://doi.org/10.1016/j.bbabio.2010.02.015.
Silkin, Y., Oyedotun, K. S., and Lemire, B. D. (2007) The role of Sdh4p Tyr-89 in ubiquinone reduction by the Saccharomyces cerevisiae succinate dehydrogenase, Biochim. Biophys. Acta, 1767, 143-150, https://doi.org/10.1016/j.bbabio.2006.11.017.
Tran, Q. M., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2006) The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b, J. Biol. Chem., 281, 32310-32317, https://doi.org/10.1074/jbc.M607476200.
Tran, Q. M., Fong, C., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2012) Out of plane distortions of the heme b of Escherichia coli succinate dehydrogenase, PLoS One, 7, e32641, https://doi.org/10.1371/journal.pone.0032641.
Maklashina, E., Rothery, R. A., Weiner, J. H., and Cecchini, G. (2001) Retention of heme in axial ligand mutants of succinate-ubiquinone oxidoreductase (complex II) from Escherichia coli, J. Biol. Chem., 276, 18968-18976, https://doi.org/10.1074/jbc.M011270200.
Gu, L. Q., Yu, L., and Yu, C. A. (1990) Effect of substituents of the benzoquinone ring on electron-transfer activities of ubiquinone derivatives, Biochim. Biophys. Acta, 1015, 482-492, https://doi.org/10.1016/0005-2728(90)90082-f.
Acknowledgments
I dedicate this review to my mentor Andrei Vinogradov. He devoted a large portion of his scientific life to study mitochondrial complex II, which he often called “my first love in science”. I also wish to thank Gary Cecchini for reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares no conflicts of interest. This article does not contain description of studies with the involvement of humans or animal subjects.
Rights and permissions
About this article
Cite this article
Maklashina, E. Structural Insight into Evolution of the Quinone Binding Site in Complex II. Biochemistry Moscow 87, 752–761 (2022). https://doi.org/10.1134/S0006297922080077
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297922080077




