Abstract
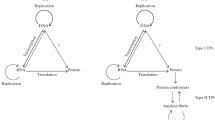
The review considers the reasons and consequences of post-transcriptional tyrosine substitutions for cysteine residues. Main attention is paid to the Tyr/Cys substitutions that arise during gene expression in bacterial systems at the stage of protein translation as a result of misrecognition of the similar mRNA codons. Notably, translation errors generally occur relatively rarely – from 10–4 to 10–3 errors per codon for E. coli cells, but in some cases the error rate increases significantly. For example, this is typical for certain pairs of codons, when the culture conditions change or in the presence of antibiotics. Thus, with overproduction of the recombinant human alpha-synuclein in E. coli cells, the content of the mutant form with the replacement of Tyr136 (UAC codon) with a cysteine residue (UGC codon) can reach 50%. Possible reasons for the increased production of alpha-synuclein with the Tyr136Cys substitution are considered, as well as consequences of the presence of mutant forms in preparations of amyloidogenic proteins when studying their pathological transformation in vitro. A separate section is devoted to the Tyr/Cys substitutions occurring due to mRNA editing by adenosine deaminases, which is typical for eukaryotic organisms, and the possible role of this process in the amyloid transformation of proteins associated with neurodegenerative diseases.


Similar content being viewed by others
References
Nishikura, K. (2010) Functions and regulation of RNA editing by ADAR deaminases, Annu. Rev. Biochem., 79, 321-349, https://doi.org/10.1146/annurev-biochem-060208-105251.
Chen, C. X., Cho, D. S., Wang, Q., Lai, F., Carter, K. C., et al. (2000) A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains, RNA, 6, 755-767, https://doi.org/10.1017/s1355838200000170.
Sinigaglia, K., Wiatrek, D., Khan, A., Michalik, D., Sambrani, N., et al. (2019) ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep, Biochim. Biophys. Acta, 1862, 356-369, https://doi.org/10.1016/j.bbagrm.2018.10.011.
Roth, S. H., Danan-Gotthold, M., Ben-Izhak, M., Rechavi, G., Cohen, C. J., et al. (2018) Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus, Cell Rep., 23, 50-57, https://doi.org/10.1016/j.celrep.2018.03.036.
Silvestris, D. A., Picardi, E., Cesarini, V., Fosso, B., Mangraviti, N., et al. (2019) Dynamic inosinome profiles reveal novel patient stratification and gender-specific differences in glioblastoma, Genome Biol., 20, 33, https://doi.org/10.1186/s13059-019-1647-x.
Costa Cruz, P. H., and Kawahara, Y. (2021) RNA Editing in Neurological and Neurodegenerative Disorders, in RNA Editing (Picardi, E., and Pesole, G., eds.) Springer US, New York, NY, pp. 309-330, https://doi.org/10.1007/978-1-0716-0787-9_18.
Parker, J. (1989) Errors and alternatives in reading the universal genetic code, Microbiol. Rev., 53, 273-298, https://doi.org/10.1128/mr.53.3.273-298.
Loftfield, R. B., and Vanderjagt, D. (1972) The frequency of errors in protein biosynthesis, Biochem. J., 128, 1353-1356, https://doi.org/10.1042/bj1281353.
Khazaie, K., Buchanan, J. H., and Rosenberger, R. F. (1984) The accuracy of Qbeta RNA translation. 1. Errors during the synthesis of Qbeta proteins by intact Escherichia coli cells, Eur. J. Biochem., 144, 485-489, https://doi.org/10.1111/j.1432-1033.1984.tb08491.x.
Kramer, E. B., and Farabaugh, P. J. (2007) The frequency of translational misreading errors in E. coli is largely determined by tRNA competition, RNA, 13, 87-96, https://doi.org/10.1261/rna.294907.
Wohlgemuth, I., Garofalo, R., Samatova, E., Günenç, A. N., Lenz, C., et al. (2021) Translation error clusters induced by aminoglycoside antibiotics, Nat. Commun., 12, 1830, https://doi.org/10.1038/s41467-021-21942-6.
Zhang, J., Pavlov, M. Y., and Ehrenberg, M. (2018) Accuracy of genetic code translation and its orthogonal corruption by aminoglycosides and Mg2+ ions, Nucleic Acids Res., 46, 1362-1374, https://doi.org/10.1093/nar/gkx1256.
Garofalo, R., Wohlgemuth, I., Pearson, M., Lenz, C., Urlaub, H., et al. (2019) Broad range of missense error frequencies in cellular proteins, Nucleic Acids Res., 47, 2932-2945, https://doi.org/10.1093/nar/gky1319.
McNulty, D. E., Claffee, B. A., Huddleston, M. J., Porter, M. L., Cavnar, K. M., et al. (2003) Mistranslational errors associated with the rare arginine codon CGG in Escherichia coli, Protein Express. Purif., 27, 365-374, https://doi.org/10.1016/s1046-5928(02)00610-1.
Calderone, T. L., Stevens, R. D., and Oas, T. G. (1996) High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli, J. Mol. Biol., 262, 407-412, https://doi.org/10.1006/jmbi.1996.0524.
Huang, Y., O’Mara, B., Conover, M., Ludwig, R., Fu, J., et al. (2012) Glycine to glutamic acid misincorporation observed in a recombinant protein expressed by Escherichia coli cells, Protein Sci., 21, 625-632, https://doi.org/10.1002/pro.2046.
Liu, Y., Sharp, J. S., Do, D. H.-T., Kahn, R. A., Schwalbe, H., et al. (2017) Mistakes in translation: Reflections on mechanism, PLoS One, 12, e0180566, https://doi.org/10.1371/journal.pone.0180566.
Zimmerman, S. M., Kon, Y., Hauke, A. C., Ruiz, B. Y., Fields, S., et al. (2018) Conditional accumulation of toxic tRNAs to cause amino acid misincorporation, Nucleic Acids Res., 46, 7831-7843, https://doi.org/10.1093/nar/gky623.
Kramer, E. B., Vallabhaneni, H., Mayer, L. M., and Farabaugh, P. J. (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae, RNA, 16, 1797-1808, https://doi.org/10.1261/rna.2201210.
Rice, J. B., Seyer, J. J., and Reeve, J. N. (1986) Identification of sites of cysteine misincorporation during in vivo synthesis of bacteriophage T7 0.3 protein, Biochim. Biophys. Acta, 867, 57-66, https://doi.org/10.1016/0167-4781(86)90029-1.
Masuda, M., Dohmae, N., Nonaka, T., Oikawa, T., Hisanaga, S., et al. (2006) Cysteine misincorporation in bacterially expressed human α-synuclein, FEBS Lett., 580, 1775-1779, https://doi.org/10.1016/j.febslet.2006.02.032.
Barinova, K. V., Kuravsky, M. L., Arutyunyan, A. M., Serebryakova, M. V., Schmalhausen, E. V., et al. (2017) Dimerization of Tyr136Cys alpha-synuclein prevents amyloid transformation of wild type alpha-synuclein, Int. J. Biological Macromol., 96, 35-43, https://doi.org/10.1016/j.ijbiomac.2016.12.011.
Kim, U., Wang, Y., Sanford, T., Zeng, Y., and Nishikura, K. (1994) Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing, Proc. Natl. Acad. Sci. USA, 91, 11457-11461, https://doi.org/10.1073/pnas.91.24.11457.
Kim, U., and Nishikura, K. (1993) Double-stranded RNA adenosine deaminase as a potential mammalian RNA editing factor, Semin. Cell Biol., 4, 285-293, https://doi.org/10.1006/scel.1993.1034.
Maas, S., Melcher, T., and Seeburg, P. H. (1997) Mammalian RNA-dependent deaminases and edited mRNAs, Curr. Opin. Cell Biol., 9, 343-349, https://doi.org/10.1016/S0955-0674(97)80006-3.
Yuting, K., Ding, D., and Iizasa, H. (2021) Adenosine-to-Inosine RNA Editing Enzyme ADAR and microRNAs, Methods Mol. Biol., 2181, 83-95, https://doi.org/10.1007/978-1-0716-0787-9_6.
Mallela, A., and Nishikura, K. (2012) A-to-I editing of protein coding and noncoding RNAs, Crit. Rev. Biochem. Mol. Biol., 47, 493-501, https://doi.org/10.3109/10409238.2012.714350.
Kliuchnikova, A. A., Kuznetsova, K. G., and Moshkovskii, S. A. (2016) ADAR-mediated messenger RNA editing: Analysis at the proteome level, Biochemistry (Moscow), Suppl. Series B Biomed. Chem., 11, 32-42, https://doi.org/10.18097/PBMC20166205510.
Maas, S., Kawahara, Y., Tamburro, K. M., and Nishikura, K. (2006) A-to-I RNA editing and human disease, RNA Biol., 3, 1-9, https://doi.org/10.4161/rna.3.1.2495.
Gaisler-Salomon, I., Kravitz, E., Feiler, Y., Safran, M., Biegon, A., et al. (2014) Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease, Neurobiol. Aging, 35, 1785-1791, https://doi.org/10.1016/j.neurobiolaging.2014.02.018.
Akbarian, S., Smith, M. A., and Jones, E. G. (1995) Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia, Brain Res., 699, 297-304, https://doi.org/10.1016/0006-8993(95)00922-D.
Hosaka, T., Tsuji, H., and Kwak, S. (2021) RNA editing: A new therapeutic target in amyotrophic lateral sclerosis and other neurological diseases, Int. J. Mol. Sci., 22, 10958, https://doi.org/10.3390/ijms222010958.
Khermesh, K., D’Erchia, A. M., Barak, M., Annese, A., Wachtel, C., et al. (2016) Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease, RNA, 22, 290-302, https://doi.org/10.1261/rna.054627.115.
Lo Giudice, C., Tangaro, M. A., Pesole, G., and Picardi, E. (2020) Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal, Nat. Protocols, 15, 1098-1131, https://doi.org/10.1038/s41596-019-0279-7.
Mansi, L., Tangaro, M. A., Lo Giudice, C., Flati, T., Kopel, E., et al. (2021) REDIportal: Millions of novel A-to-I RNA editing events from thousands of RNAseq experiments, Nucleic Acids Res., 49, D1012-D1019, https://doi.org/10.1093/nar/gkaa916.
Hatos, A., Hajdu-Soltész, B., Monzon, A. M., Palopoli, N., Álvarez, L., et al. (2019) DisProt: Intrinsic protein disorder annotation in 2020, Nucleic Acids Res., 48, D269-D276, https://doi.org/10.1093/nar/gkz975.
Ramaswami, G., and Li, J. B. (2014) RADAR: A rigorously annotated database of A-to-I RNA editing, Nucleic Acids Res., 42, D109-D113, https://doi.org/10.1093/nar/gkt996.
Picardi, E., D’Erchia, A. M., Lo Giudice, C., and Pesole, G. (2017) REDIportal: A comprehensive database of A-to-I RNA editing events in humans, Nucleic Acids Res., 45, D750-D757, https://doi.org/10.1093/nar/gkw767.
Pozdyshev, D. V., Melnikova, A. K., Barinova, K. V., Schmalhausen, E. V., and Muronetz, V. I. (2020) Differences in the synthesis of recombinant α-synuclein in pro-and eukaryotic organisms: Possibility of Tyr136Cys substitution, Curr. Top. Peptide Prot. Res., 21, 75-81.
Feughelman, M., and Willis, B. K. (2000) Thiol-disulfide interchange a potential key to conformational change associated with amyloid fibril formation, J. Theor. Biol., 206, 313-315, https://doi.org/10.1006/jtbi.2000.2112.
Li, Y., Yan, J., Zhang, X., and Huang, K. (2013) Disulfide bonds in amyloidogenesis diseases related proteins, Proteins, 81, 1862-1873, https://doi.org/10.1002/prot.24338.
Maiti, N. R., and Surewicz, W. K. (2001) The role of disulfide bridge in the folding and stability of the recombinant human prion protein, J. Biol. Chem., 276, 2427-2431, https://doi.org/10.1074/jbc.M007862200.
Lee, S., and Eisenberg, D. (2003) Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process, Nat. Struct. Biol., 10, 725-730, https://doi.org/10.1038/nsb961.
Hosszu, L. L. P., Trevitt, C. R., Jones, S., Batchelor, M., Scott, D. J., et al. (2009) Conformational properties of beta-PrP, J. Biol. Chem., 284, 21981-21990, https://doi.org/10.1074/jbc.M809173200.
Welker, E., Wedemeyer, W. J., and Scheraga, H. A. (2001) A role for intermolecular disulfide bonds in prion diseases? Proc. Natl. Acad. Sci. USA, 98, 4334-4336, https://doi.org/10.1073/pnas.071066598.
Mehlhorn, I., Groth, D., Stöckel, J., Moffat, B., Reilly, D., et al. (1996) High-level expression and characterization of a purified 142-residue polypeptide of the prion protein, Biochemistry, 35, 5528-5537, https://doi.org/10.1021/bi952965e.
Suk, J.-E., Lokappa, S. B., and Ulmer, T. S. (2010) The clustering and spatial arrangement of beta-sheet sequence, but not order, govern alpha-synuclein fibrillogenesis, Biochemistry, 49, 1533-1540, https://doi.org/10.1021/bi901753h.
Zhou, W., and Freed, C. R. (2004) Tyrosine-to-cysteine modification of human alpha-synuclein enhances protein aggregation and cellular toxicity, J. Biol. Chem., 279, 10128-10135, https://doi.org/10.1074/jbc.M307563200.
Krishnan, R., and Lindquist, S. L. (2005) Structural insights into a yeast prion illuminate nucleation and strain diversity, Nature, 435, 765-772, https://doi.org/10.1038/nature03679.
Hong, D.-P., Xiong, W., Chang, J.-Y., and Jiang, C. (2011) The role of the C-terminus of human α-synuclein: Intra-disulfide bonds between the C-terminus and other regions stabilize non-fibrillar monomeric isomers, FEBS Lett., 585, 561-566, https://doi.org/10.1016/j.febslet.2011.01.009.
Funding
This work was financially supported by the Russian Foundation for Basic Research (project no. 19-04-00421).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflicts of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Muronetz, V.I., Pozdyshev, D.V., Medvedeva, M.V. et al. Potential Effect of Post-Transcriptional Substitutions of Tyrosine for Cysteine Residues on Transformation of Amyloidogenic Proteins. Biochemistry Moscow 87, 170–178 (2022). https://doi.org/10.1134/S0006297922020080
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297922020080