Abstract
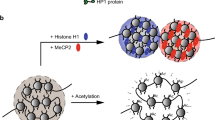
Functional compartmentalization of the cell nucleus plays an important role in the regulation of genome activity by providing accumulation of enzymes and auxiliary factors in the reaction centers, such as transcription factories, Cajal bodies, speckles, etc. The mechanisms behind the nucleus functional compartmentalization are still poorly understood. There are reasons to believe that the key role in the nucleus compartmentalization belongs to the process of liquid–liquid phase separation. In this brief review, we analyze results of experimental studies demonstrating that liquid–liquid phase separation not only governs functional compartmentalization of the cell nucleus but also contributes to the formation of the 3D genomic architecture.


Similar content being viewed by others
Abbreviations
- IC:
-
interchromatin compartment
- IDR:
-
intrinsically disordered region
REFERENCES
Hancock, R. (2004) Internal organisation of the nucleus: assembly of compartments by macromolecular crowding and the nuclear matrix model, Biol. Cell, 96, 595-601.
Hancock, R. (2004) A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus, J. Struct. Biol., 146, 281-290, doi: 10.1016/j.jsb.2003.12.008.
Hancock, R. (2018) Crowding, entropic forces, and confinement: crucial factors for structures and functions in the cell nucleus, Biochemistry (Moscow), 83, 326-337, doi: 10.1134/S0006297918040041.
Marenduzzo, D., Finan, K., and Cook, P. R. (2006) The depletion attraction: an underappreciated force driving cellular organization, J. Cell. Biol., 175, 681-686, doi: 10.1083/jcb.200609066.
Razin, S. V., Gavrilov, A. A., Pichugin, A., Lipinski, M., Iarovaia, O. V., and Vassetzky, Y. S. (2011) Transcription factories in the context of the nuclear and genome organization, Nucleic Acids Res., 39, 9085-9092, doi: 10.1093/nar/gkr683.
Erdel, F., and Rippe, K. (2018) Formation of chromatin subcompartments by phase separation, Biophys. J., 114, 2262-2270, doi: 10.1016/j.bpj.2018.03.011.
Boeynaems, S., Alberti, S., Fawzi, N. L., Mittag, T., Polymenidou, M., Rousseau, F., Schymkowitz, J., Shorter, J., Wolozin, B., Van Den Bosch, L., Tompa, P., and Fuxreiter, M. (2018) Protein phase separation: a new phase in cell biology, Trends Cell. Biol., 28, 420-435, doi: 10.1016/j.tcb.2018.02.004.
Uversky, V. N. (2017) Protein intrinsic disorder-based liquid–liquid phase transitions in biological systems: complex coacervates and membrane-less organelles, Adv. Coll. Interface Sci., 239, 97-114, doi: 10.1016/j.cis.2016.05.012.
Meng, F., Na, I., Kurgan, L., and Uversky, V. N. (2015) Compartmentalization and functionality of nuclear disorder: intrinsic disorder and protein–protein interactions in intra-nuclear compartments, Intern. J. Mol. Sci., 17, doi: 10.3390/ijms17010024.
Darling, A. L., Liu, Y., Oldfield, C. J., and Uversky, V. N. (2018) Intrinsically disordered proteome of human membrane-less organelles, Proteomics, 18, e1700193, doi: 10.1002/pmic.201700193.
Uversky, V. N. (2017) Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder, Curr. Opin. Struct. Biol., 44, 18-30, doi: 10.1016/j.sbi.2016.10.015.
Turner, A. L., Watson, M., Wilkins, O. G., Cato, L., Travers, A., Thomas, J. O., and Stott, K. (2018) Highly disordered histone H1–DNA model complexes and their condensates, Proc. Natl. Acad. Sci. USA, 115, 11964-11969, doi: 10.1073/pnas.1805943115.
Larson, A. G., Elnatan, D., Keenen, M. M., Trnka, M. J., Johnston, J. B., Burlingame, A. L., Agard, D. A., Redding, S., and Narlikar, G. J. (2017) Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin, Nature, 547, 236-240, doi: 10.1038/nature22822.
Tatavosian, R., Kent, S., Brown, K., Yao, T., Duc, H. N., Huynh, T. N., Zhen, C. Y., Ma, B., Wang, H., and Ren, X. (2019) Nuclear condensates of the polycomb protein chromobox 2 (CBX2) assemble through phase separation, J. Biol. Chem., 294, 1451-1463, doi: 10.1074/jbc.RA118.006620.
Boehning, M., Dugast-Darzacq, C., Rankovic, M., Hansen, A. S., Yu, T., Marie-Nelly, H., McSwiggen, D. T., Kokic, G., Dailey, G. M., Cramer, P., Darzacq, X., and Zweckstetter, M. (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation, Nat. Struct. Mol. Biol., 25, 833-840, doi: 10.1038/s41594-018-0112-y.
Nagulapalli, M., Maji, S., Dwivedi, N., Dahiya, P., and Thakur, J. K. (2016) Evolution of disorder in mediator complex and its functional relevance, Nucleic Acids Res., 44, 1591-1612, doi: 10.1093/nar/gkv1135.
Alberti, S., Gladfelter, A., and Mittag, T. (2019) Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates, Cell, 176, 419-434, doi: 10.1016/j.cell.2018.12.035.
Sabari, B. R., Dall’Agnese, A., Boija, A., Klein, I. A., Coffey, E. L. et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control, Science, 361, doi: 10.1126/science.aar3958.
Cho, W. K., Spille, J. H., Hecht, M., Lee, C., Li, C., Grube, V., and Cisse, I. I. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates, Science, 361, 412-415, doi: 10.1126/science. aar4199.
Hernandez-Verdun, D. (2006) The nucleolus: a model for the organization of nuclear functions, Histochem. Cell Biol., 126, 135-148, doi: 10.1007/s00418-006-0212-3.
Yao, R. W., Xu, G., Wang, Y., Shan, L., Luan, P. F., Wang, Y., Wu, M., Yang, L. Z., Xing, Y. H., Yang, L., and Chen, L. L. (2019) Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus, Mol. Cell, 76, 767-783, e711, doi: 10.1016/j.molcel.2019.08.014.
Correll, C. C., Bartek, J., and Dundr, M. (2019) The nucleolus: a multiphase condensate balancing ribosome synthesis and translational capacity in health, aging and ribosomopathies, Cells, 8, doi: 10.3390/cells8080869.
Ishov, A. M., Sotnikov, A. G., Negorev, D., Vladimirova, O. V., Neff, N., Kamitani, T., Yeh, E. T., Strauss, J. F. 3rd, and Maul, G. G. (1999) PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1, J. Cell. Biol., 147, 221-234, doi: 10.1083/jcb.147.2.221.
Lallemand-Breitenbach, V., and de Thé, H. (2010) PML nuclear bodies, Cold Spring Harb. Perspect. Biol., 2, a000661.
Yamazaki, T., Nakagawa, S., and Hirose, T. (2020) Architectural RNAs for membraneless nuclear body formation, Cold Spring Harb. Symp. Quant. Biol., doi: 10.1101/sqb.2019.84.039404.
Fox, A. H., and Lamond, A. I. (2010) Paraspeckles, Cold Spring Harb. Perspect. Biol., 2, a000687, doi: 10.1101/cshperspect.a000687.
Fox, A. H., Nakagawa, S., Hirose, T., and Bond, C. S. (2018) Paraspeckles: where long noncoding RNA meets phase separation, Trends Biochem. Sci., 43, 124-135, doi: 10.1016/j.tibs.2017.12.001.
Shin, Y., Berry, J., Pannucci, N., Haataja, M. P., Toettcher, J. E., and Brangwynne, C. P. (2017) Spatiotemporal control of intracellular phase transitions using light-activated optodroplets, Cell, 168, 159-171, doi: 10.1016/j.cell.2016.11.054.
Zhou, J., Fan, J. Y., Rangasamy, D., and Tremethick, D. J. (2007) The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression, Nat. Struct. Mol. Biol., 14, 1070-1076.
Kalashnikova, A. A., Porter-Goff, M. E., Muthurajan, U. M., Luger, K., and Hansen, J. C. (2013) The role of the nucleosome acidic patch in modulating higher order chromatin structure, J. R. Soc. Interface, 10, 20121022, doi: 10.1098/rsif.2012.1022.
Sinha, D., and Shogren-Knaak, M. A. (2010) Role of direct interactions between the histone H4 tail and the H2A core in long range nucleosome contacts, J. Biol. Chem., 285, 16572-16581, doi: 10.1074/jbc.M109.091298.
Pepenella, S., Murphy, K. J., and Hayes, J. J. (2014) Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure, Chromosoma, 123, 3-13, doi: 10.1007/s00412-013-0435-8.
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution, Nature, 389, 251-260, doi: 10.1038/38444.
Schalch, T., Duda, S., Sargent, D. F., and Richmond, T. J. (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre, Nature, 436, 138-141, doi: 10.1038/nature03686.
Chodaparambil, J. V., Barbera, A. J., Lu, X., Kaye, K. M., Hansen, J. C., and Luger, K. (2007) A charged and contoured surface on the nucleosome regulates chromatin compaction, Nat. Struct. Mol. Biol., 14, 1105-1107, doi: 10.1038/nsmb1334.
Chen, Q., Yang, R., Korolev, N., Liu, C. F., and Nordenskiold, L. (2017) Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail-H2A acidic patch interaction, J. Mol. Biol., 429, 2075-2092, doi: 10.1016/j.jmb.2017.03.016.
Gibson, B. A., Doolittle, L. K., Schneider, M. W. G., Jensen, L. E., Gamarra, N., Henry, L., Gerlich, D. W., Redding, S., and Rosen, M. K. (2019) Organization of chromatin by intrinsic and regulated phase separation, Cell, 179, 470-484, doi: 10.1016/j.cell.2019.08.037.
Shakya, A., Park, S., Rana, N., and King, J. T. (2020) Liquid–liquid phase separation of histone proteins in cells: role in chromatin organization, Biophys. J., 118, 753-764, doi: 10.1016/j.bpj.2019.12.022.
Strom, A. R., Emelyanov, A. V., Mir, M., Fyodorov, D. V., Darzacq, X., and Karpen, G. H. (2017) Phase separation drives heterochromatin domain formation, Nature, 547, 241-245, doi: 10.1038/nature22989.
Strom, A. R., and Brangwynne, C. P. (2019) The liquid nucleome – phase transitions in the nucleus at a glance, J. Cell. Sci., 132, jcs235093, doi: 10.1242/jcs.235093.
Plys, A. J., Davis, C. P., Kim, J., Rizki, G., Keenen, M. M., Marr, S. K., and Kingston, R. E. (2019) Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2, Genes Dev., 33, 799-813, doi: 10.1101/gad.326488.119.
Peng, A., and Weber, S. C. (2019) Evidence for and against liquid–liquid phase separation in the nucleus, Noncoding RNA, 5, doi: 10.3390/ncrna5040050.
Boija, A., Klein, I. A., Sabari, B. R., Dall’Agnese, A., Coffey, E. L., Zamudio, A. V., Li, C. H., Shrinivas, K., Manteiga, J. C., Hannett, N. M., Abraham, B. J., Afeyan, L. K., Guo, Y. E., Rimel, J. K., Fant, C. B., Schuijers, J., Lee, T. I., Taatjes, D. J., and Young, R. A. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains, Cell, 175, 1842-1855, doi: 10.1016/j.cell.2018.10.042.
Tarczewska, A., and Greb-Markiewicz, B. (2019) The significance of the intrinsically disordered regions for the functions of the bHLH transcription factors, Intern. J. Mol. Sci., 20, doi: 10.3390/ijms20215306.
Cremer, T., and Cremer, M. (2010) Chromosome territories, Cold Spring Harb. Perspect. Biol., 2, a003889, doi: 10.1101/cshperspect.a003889.
Cremer, T., Cremer, M., Hubner, B., Silahtaroglu, A., Hendzel, M., Lanctot, C., Strickfaden, H., and Cremer, C. (2020) The interchromatin compartment participates in the structural and functional organization of the cell nucleus, BioEssays, 42, e1900132, doi: 10.1002/bies.201900132.
Cremer, T., Cremer, M., and Cremer, C. (2018) The 4D nucleome: genome compartmentalization in an evolutionary context, Biochemistry (Moscow), 83, 313-325, doi: 10.1134/S000629791804003X.
Garcia-Jove Navarro, M., Kashida, S., Chouaib, R., Souquere, S., Pierron, G., Weil, D., and Gueroui, Z. (2019) RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates, Nat. Commun., 10, 3230, doi: 10.1038/s41467-019-11241-6.
Fay, M. M., and Anderson, P. J. (2018) The role of RNA in biological phase separations, J. Mol. Biol., 430, 4685-4701, doi: 10.1016/j.jmb.2018.05.003.
Fedoriw, A. M., Starmer, J., Yee, D., and Magnuson, T. (2012) Nucleolar association and transcriptional inhibition through 5S rDNA in mammals, PLoS Genet., 8, e1002468, doi: 10.1371/journal.pgen.1002468.
Bersaglieri, C., and Santoro, R. (2019) Genome organization in and around the nucleolus, Cells, 8, doi: 10.3390/cells8060579.
Chen, Y., Zhang, Y., Wang, Y., Zhang, L., Brinkman, E. K., Adam, S. A., Goldman, R., van Steensel, B., Ma, J., and Belmont, A. S. (2018) Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler, J. Cell. Biol., 217, 4025-4048, doi: 10.1083/jcb.201807108.
Quinodoz, S. A., Ollikainen, N., Tabak, B., Palla, A., Schmidt, J. M., Detmar, E., Lai, M. M., Shishkin, A. A., Bhat, P., Takei, Y., Trinh, V., Aznauryan, E., Russell, P., Cheng, C., Jovanovic, M., Chow, A., Cai, L., McDonel, P., Garber, M., and Guttman, M. (2018) Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus, Cell, 174, 744-757, doi: 10.1016/j.cell.2018.05.024.
Kim, J., Venkata, N. C., Hernandez Gonzalez, G. A., Khanna, N., and Belmont, A. S. (2020) Gene expression amplification by nuclear speckle association, J. Cell. Biol., 219, e201904046, doi: 10.1083/jcb.201904046.
Dundr, M. (2012) Nuclear bodies: multifunctional companions of the genome, Curr. Opin. Cell. Biol., 24, 415-422, doi: 10.1016/j.ceb.2012.03.010.
Sawyer, I. A., Sturgill, D., and Dundr, M. (2019) Membraneless nuclear organelles and the search for phases within phases, Wiley Interdiscip. Rev. RNA, 10, e1514, doi: 10.1002/wrna.1514.
Wang, Q., Sawyer, I. A., Sung, M. H., Sturgill, D., Shevtsov, S. P., Pegoraro, G., Hakim, O., Baek, S., Hager, G. L., and Dundr, M. (2016) Cajal bodies are linked to genome conformation, Nat. Commun., 7, 10966, doi: 10.1038/ncomms10966.
Carter, D. R., Eskiw, C., and Cook, P. R. (2008) Transcription factories, Biochem. Soc. Trans., 36, 585-589, doi: 10.1042/BST0360585.
Hozak, P., Hassan, A. B., Jackson, D. A., and Cook, P. R. (1993) Visualization of replication factories attached to nucleoskeleton, Cell, 73, 361-373.
Iborra, F. J., Pombo, A., Jackson, D. A., and Cook, P. R. (1996) Active RNA polymerases are localized within discrete transcription “factories” in human nuclei, J. Cell Sci., 109, 1427-1436.
Jackson, D. A., Hassan, A. B., Errington, R. J., and Cook, P. R. (1993) Visualization of focal sites of transcription within human nuclei, EMBO J., 12, 1059-1065.
Sutherland, H., and Bickmore, W. A. (2009) Transcription factories: gene expression in unions? Nat. Rev. Genet., 10, 457-466.
Cook, P. R., and Marenduzzo, D. (2018) Transcription-driven genome organization: a model for chromosome structure and the regulation of gene expression tested through simulations, Nucleic Acids Res., 46, 9895-9906, doi: 10.1093/nar/gky763.
Osborne, C. S., Chakalova, L., Brown, K. E., Carter, D., Horton, A., Debrand, E., Goyenechea, B., Mitchell, J. A., Lopes, S., Reik, W., and Fraser, P. (2004) Active genes dynamically colocalize to shared sites of ongoing transcription, Nat. Genet., 36, 1065-1071.
Osborne, C. S., Chakalova, L., Mitchell, J. A., Horton, A., Wood, A. L., Bolland, D. J., Corcoran, A. E., and Fraser, P. (2007) Myc dynamically and preferentially relocates to a transcription factory occupied by Igh, PLoS Biol., 5, e192.
Ulianov, S. V., Doronin, S. A., Khrameeva, E. E., Kos, P. I., Luzhin, A. V., Starikov, S. S., Galitsyna, A. A., Nenasheva, V. V., Ilyin, A. A., Flyamer, I. M., Mikhaleva, E. A., Logacheva, M. D., Gelfand, M. S., Chertovich, A. V., Gavrilov, A. A., Razin, S. V., and Shevelyov, Y. Y. (2019) Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila, Nat. Commun., 10, 1176, doi: 10.1038/s41467-019-09185-y.
Arnold, C. D., Gerlach, D., Stelzer, C., Boryn, L. M., Rath, M., and Stark, A. (2013) Genome-wide quantitative enhancer activity maps identified by STARR-seq, Science, 339, 1074-1077, doi: 10.1126/science.1232542.
Consortium, E. P., Bernstein, B. E., Birney, E., Dunham, I., Green, E. D., Gunter, C., and Snyder, M. (2012) An integrated encyclopedia of DNA elements in the human genome, Nature, 489, 57-74, doi: 10.1038/nature11247.
Furlong, E. E. M., and Levine, M. (2018) Developmental enhancers and chromosome topology, Science, 361, 1341-1345, doi: 10.1126/science.aau0320.
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K., and Sharp, P. A. (2017) A phase separation model for transcriptional control, Cell, 169, 13-23, doi: 10.1016/j.cell.2017.02.007.
Gurumurthy, A., Shen, Y., Gunn, E. M., and Bungert, J. (2019) Phase separation and transcription regulation: are super-enhancers and locus control regions primary sites of transcription complex assembly? BioEssays, 41, e1800164, doi: 10.1002/bies.201800164.
Arnold, P. R., Wells, A. D., and Li, X. C. (2019) Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate, Front. Cell Develop. Biol., 7, 377, doi: 10.3389/fcell.2019.00377.
Nair, S. J., Yang, L., Meluzzi, D., Oh, S., Yang, F., Friedman, M. J., Wang, S., Suter, T., Alshareedah, I., Gamliel, A., Ma, Q., Zhang, J., Hu, Y., Tan, Y., Ohgi, K. A., Jayani, R. S., Banerjee, P. R., Aggarwal, A. K., and Rosenfeld, M. G. (2019) Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly, Nat. Struct. Mol. Biol., 26, 193-203, doi: 10.1038/s41594-019-0190-5.
Hyman, A. A., Weber, C. A., and Julicher, F. (2014) Liquid–liquid phase separation in biology, Ann. Rev. Cell Dev. Biol., 30, 39-58, doi: 10.1146/annurev-cellbio-100913-013325.
Cramer, P. (2019) Organization and regulation of gene transcription, Nature, 573, 45-54, doi: 10.1038/s41586-019-1517-4.
Guo, Y. E., Manteiga, J. C., Henninger, J. E., Sabari, B. R., Dall’Agnese, A., Hannett, N. M., Spille, J. H., Afeyan, L. K., Zamudio, A. V., Shrinivas, K., Abraham, B. J., Boija, A., Decker, T. M., Rimel, J. K., Fant, C. B., Lee, T. I., Cisse, I. I., Sharp, P. A., Taatjes, D. J., and Young, R. A. (2019) Pol II phosphorylation regulates a switch between transcriptional and splicing condensates, Nature, 572, 543-548, doi: 10.1038/s41586-019-1464-0.
Erdel, F., Rademacher, A., Vlijm, R., Tunnermann, J., Frank, L., Weinmann, R., Schweigert, E., Yserentant, K., Hummert, J., Bauer, C., Schumacher, S., Al Alwash, A., Normand, C., Herten, D. P., Engelhardt, J., and Rippe, K. (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid–liquid phase separation, Mol. Cell, 78, 236-249 doi: 10.1016/j.molcel.2020.02.005.
Mir, M., Bickmore, W., Furlong, E. E. M., and Narlikar, G. (2019) Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development, 146, doi: 10.1242/dev.182766.
Funding
This work was financially supported by the Russian Science Foundation (project No. 18-14-00011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain studies with human participants or animals performed by any of the authors. The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Razin, S., Gavrilov, A. The Role of Liquid–Liquid Phase Separation in the Compartmentalization of Cell Nucleus and Spatial Genome Organization. Biochemistry Moscow 85, 643–650 (2020). https://doi.org/10.1134/S0006297920060012
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920060012