Abstract
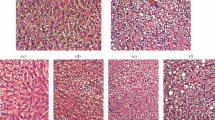
Differential expression of 30,003 genes was studied in the liver of female Wistar rats fed with isocaloric diets with the excess of fat, fructose, or cholesterol, or their combinations for 62 days using the method of whole-transcriptome pro-filing on a microchip. Relative mRNA expression levels of the Asah2, Crot, Crtc2, Fmo3, GSTA2, LOC1009122026, LOC102551184, NpY, NqO1, Prom1, Retsat, RGD1305464, Tmem104, and Whsc1 genes were also determined by RT-qPCR. All the tested diets affected differently the key metabolic pathways (KEGGs). Significant changes in the expression of steroid metabolism gene were observed in the liver of animals fed with the tested diets (except the high-fat high fructose diet). Both high-fat and high-fructose diets caused a significant decrease in the expression of squalene synthase (FDFT1 gene) responsible for the initial stage of cholesterol synthesis. On the contrary, in animals fed with the high-cholesterol diet (0.5% cholesterol), expression of the FDFT1 gene did not differ from the control group; however, these animals were characterized by changes in the expression of glucose and glycogen synthesis genes, which could lead to the suppression of glycogen synthesis and gluconeogenesis. At the same time, this group demonstrated different liver tissue morphology in comparison with the animals fed with the high-fructose high-fat diet, manifested as the presence of lipid vacuoles of a smaller size in hepatocytes. The high-fructose and high-fructose high-fat diets affected the metabolic pathways associated with intracellular protein catabolism (endocytosis, phagocytosis, proteasomal degradation, protein processing in the endoplasmic reticulum), tight junctions and intercellular contacts, adhesion molecules, and intracellular RNA transport. Rats fed with the high-fructose high-fat or high-cholesterol diets demonstrated consistent changes in the expression of the Crot, Prom1, and RGD1305464 genes, which reflected a coordinated shift in the regulation of lipid and carbohydrate metabolisms.
Similar content being viewed by others
Abbreviations
- AIN93M:
-
93M diet of the American Institute of Nutrition
- Asah2 :
-
N-acylsphingosine amidohydrolase 2 gene (ceramidase)
- Crot :
-
carnitine octanoyltransferase gene
- Crtc2 :
-
CREB-regulated transcription coactivator 2
- FAM:
-
carboxyfluorescein
- FDFT1 :
-
farnesyl-diphosphate farnesyltransferase 1 (squalene synthase) gene
- Fmo3 :
-
flavin-containing monooxygenase 3 gene
- GSTA2 :
-
glutathione-S-transferase alpha 2 gene
- HCD:
-
high-cholesterol diet
- HCHFrD:
-
high-cholesterol high-fructose diet
- HFaD:
-
high-fat diet
- HFaHFrD:
-
high-fat high-fructose diet
- HFrD:
-
high-fructose diet
- Inhbb :
-
inhibin beta B chain (activin β-subunit) gene
- LOC1009122026 :
-
1009122026 gene
- LOC102551184 :
-
102551184 gene
- MS:
-
metabolic syndrome
- NpY :
-
neuropeptide Y gene
- NqO1 :
-
NAD(P)H dehydrogenase, (quinone 1) gene
- Prom1 :
-
prominin 1 gene
- Retsat :
-
retinol saturase gene
- RGD1305464 (Sept14):
-
GTP-binding cytoskeletal protein 1305464 or SEPT14 gene
- RT-qPCR:
-
reverse transcription/quantitative polymerase chain reaction
- SSD:
-
semisynthetic diet
- TGF-β:
-
tumor growth factor beta
- Tmem104 :
-
transmembrane protein 104 gene
- Ugt2b37 :
-
uridine diphosphate glycosyltransferase 2 family, member b17 gene
- Whsc1 :
-
Wolf-Hirschhorn syndrome candidate 1 gene
References
Woods, S. C., Seeley, R. J., Rushing, P. A., D’Alessio, D., and Tso, P. (2003) A controlled high-fat diet induces an obese syndrome in rats, J. Nutr., 133, 1081–1087; doi: https://doi.org/10.1093/jn/133.4.1081.
Rask-Madsen, C., and Kahn, C. (2012) Tissue-specific insulin signaling, metabolic syndrome and cardiovascular disease, Arterioscler. Thromb. Vasc. Biol., 32, 2052–2059; doi: https://doi.org/10.1161/ATVBAHA.111.241919.
Dietrich, P., and Hellerbrand, C. (2014) Non-alcoholic fatty liver disease, obesity and the metabolic syndrome, Best Pract. Res. Clin. Gastroenterol., 28, 637–653; doi: https://doi.org/10.1016/j.bpg.2014.07.00.
Catrysse, L., and van Loo, G. (2017) Inflammation and the metabolic syndrome: the tissue-specific functions of NF-κB trends, Cell Biol., 27, 417–429; doi: https://doi.org/10.1016/j.tcb.2017.01.006.
Wong, S. K., Chin, K.-Y., Suhaimi, F. H., Fairus, A., and Ima-Nirwana, S. (2016) Animal models of metabolic syndrome: a review, Nutr. Metab. (Lond), 13, 65–77; doi: https://doi.org/10.1186/s12986-016-0123-9.
Kim, Y., and Park, T. (2010) DNA microarrays to define and search for genes associated with obesity, Biotechnol. J., 5, 99–112; doi: https://doi.org/10.1002/biot.200900228.
Soltis, A. R., Kennedy, N. J., Xin, X., Zhou, F., Ficarro, S. B., Yap, Y. S., Matthews, B. J., Lauffenburger, D. A., White, F. M., Marto, J. A., Davis, R. J., and Fraenkel, E. (2017) Hepatic dysfunction caused by consumption of a high-fat diet, Cell Rep., 21, 3317–3328; doi: https://doi.org/10.1016/j.celrep.2017.11.059.
Softic, S., Gupta, M. K., Wang, G. X., Fujisaka, S., O’Neill, B. T., Rao, T. N., Willoughby, J., Harbison, C., Fitzgerald, K., Ilkayeva, O., Newgard, C. B., Cohen, D. E., and Kahn, C. R. (2017) Divergent effects of glucose and fructose on hepatic lipogenesis and insulin signaling, J. Clin. Invest., 127, 4059–4074; doi: https://doi.org/10.1172/JCI94585.
Kirpich, I. A., Gobejishvili, L. N., Bon Homme, M., Waigel, S., Cave, M., Arteel, G., Barve, S. S., McClain, C. J., and Deaciuc, I. V. (2011) Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease, J. Nutr. Biochem., 22, 38–45; doi: https://doi.org/10.1016/j.jnutbio.2009.11.009
Kim, S., Sohn, I., Ahn, J. I., Lee, K. H., Lee, Y. S., and Lee, Y. S. (2004) Hepatic gene expression profiles in a long-term high-fat diet-induced obesity mouse model, Gene, 340, 99–109; doi: https://doi.org/10.1016/j.gene.2004.06.015.
Hasebe, T., Tanaka, H., Sawada, K., Nakajima, S., Ohtake, T., Fujiya, M., and Kohgo, Y. (2017) Bone morphogenetic protein-binding endothelial regulator of liver sinusoidal endothelial cells induces iron overload in a fatty liver mouse model, J. Gastroenterol., 52, 341–351; doi: https://doi.org/10.1007/s00535-016-1237-6.
Liu, Y., Cheng, F., Luo, Y. X., Hu, P., Ren, H., and Peng, M. L. (2017) The role of cytochrome P450 in nonalcoholic fatty liver induced by high-fat diet: a gene expression profile analysis, Zhonghua Gan Zang Bing Za Zhi., 25, 285–290; doi: https://doi.org/10.3760/cma.j.issn.1007-3418.2017.04.010.
Kim, J., Kwon, E. Y., Park, S., Kim, J. R., Choi, S. W., Choi, M. S., and Kim, S. J. (2016) Integrative systems analysis of diet-induced obesity identified a critical transition in the transcriptomes of the murine liver and epididymal white adipose tissue, Int. J. Obes. (Lond.), 40, 338–345; doi: https://doi.org/10.1038/ijo.2015.147.
Patsouris, D., Reddy, J. K., Muller, M., and Kersten, S. (2006) Peroxisome proliferator-activated receptor α mediates the effects of high-fat diet on hepatic gene expression, Endocrinology, 147, 1508–1516; doi: https://doi.org/10.1210/en.2005-1132.
Holvoet, P., Rull, A., Garcia-Heredia, A., Lopez-Sanroma, S., Geeraert, B., Joven, J., and Camps, J. (2015) Stevia-derived compounds attenuate the toxic effects of ectopic lipid accumulation in the liver of obese mice: a transcriptomic and metabolomic study, Food Chem. Toxicol., 77, 22–33; doi: https://doi.org/10.1016/j.fct.2014.12.017.
Chartoumpekis, D. V., Ziros, P. G., Zaravinos, A., Iskrenova, R. P., Psyrogiannis, A. I., Kyriazopoulou, V. E., Sykiotis, G. P., and Habeos, I. G. (2013) Hepatic gene expression profiling in Nrf2 knockout mice after long-term high-fat diet-induced obesity, Oxid. Med. Cell Longev., 2013, 340731; doi: https://doi.org/10.1155/2013/340731.
Knebel, B., Hartwig, S., Jacob, S., Kettel, U., Schiller, M., Passlack, W., Koellmer, C., Lehr, S., Muller-Wieland, D., and Kotzka, J. (2018) Inactivation of SREBP-1a phosphorylation prevents fatty liver disease in mice: identification of related signaling pathways by gene expression profiles in liver and proteomes of peroxisomes, Int. J. Mol. Sci., 19, E980; doi: https://doi.org/10.3390/ijms19040980.
Guide for the Care and Use of Laboratory Animals, 8th Edn., Committee for the Update of the Guide for the Care and Use of Laboratory Animals, Institute for Laboratory Animal Research (ILAR), Division on Earth and Life Studies (DELS), National Research Council of the National Academies (2011) The National Academies Press, Washington.
Reeves, P. G., Nielsen, F. H., and Fahey, G. C. (1993) AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet, J. Nutr., 123, 1939–1951; doi: https://doi.org/10.1093/jn/123.11.1939.
Agilent total RNA isolation mini kit. Protocol, 5th Edn. (2015) URL: http://www.agilent.com/cs/library/usermanuals/Public/5188_2710_A1.pdf.
One-color microarray-based gene expression analysis (low input quick Amp labeling), v. 6.9.1 (2015) URL: http://www.agilent.com/cs/library/usermanuals/Public/G4140-90040_GeneExpression_OneColor_6.9.pdf.
Roskin, G. I., and Levinson, L. B. (1957) Microscopy Techniques [in Russian], Sovetskaya Nauka, Moscow.
Benjamini, Y., and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. R. Statist. Soc. B, 57, 289–300; doi: https://doi.org/10.2307/2346101.
Brown, C. W., Houston-Hawkins, D. E., Woodruff, T. K., and Matzuk, M. M. (2000) Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions, Nat. Genet., 25, 453–457; doi: https://doi.org/10.1038/78161.
Beaulieu, M., Levesque, E., Tchernof, A., Beatty, B. G., Belanger, A., and Hum, D. W. (1997) Chromosomal localization, structure, and regulation of the UGT2B17 gene, encoding a C19 steroid metabolizing enzyme, DNA Cell Biol., 16, 1143–1154; doi: https://doi.org/10.1089/dna.1997.16.1143.
Capel, F., Rolland-Valognes, G., Dacquet, C., Brun, M., Lonchampt, M., Ktorza, A., Lockhart, B., and Galizzi, J. P. (2013) Analysis of sterol-regulatory element-binding protein 1c target genes in mouse liver during aging and high-fat diet, J. Nutrigenet. Nutrigenom., 6, 107–122; doi: https://doi.org/10.1159/000350751.
Choi, J. Y., McGregor, R. A., Kwon, E. Y., Kim, Y. J., Han, Y., Park, J. H., Lee, K. W., Kim, S. J., Kim, J., Yun, J. W., and Choi, M. S. (2016) The metabolic response to a high-fat diet reveals obesity-prone and -resistant phenotypes in mice with distinct mRNA-seq transcriptome profiles, Int. J. Obes. (Lond.), 40, 1452–1460; doi: https://doi.org/10.1038/ijo.2016.70.
Do, R., Kiss, R. S., Gaudet, D., and Engert, J. C. (2009) Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway, Clin. Genet., 75, 19–29; doi: https://doi.org/10.1111/j.1399-0004.2008.01099.x.
Torrente, Y., Belicchi, M., Sampaolesi, M., Pisati, F., Meregalli, M., D’Antona, G., Tonlorenzi, R., Porretti, L., Gavina, M., Mamchaoui, K., Pellegrino, M. A., Furling, D., Mouly, V., Butler-Browne, G. S., Bottinelli, R., Cossu, G., and Bresolin, N. (2004) Human circulating AC133+ stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle, J. Clin. Invest., 114, 182–195; doi: https://doi.org/10.1172/JCI20325.
Zhu, L., Gibson, P., Currle, D. S., Tong, Y., Richardson, R. J., Bayazitov, I. T., Poppleton, H., Zakharenko, S., Ellison, D. W., and Gilbertson, R. J. (2009) Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation, Nature, 457, 603–607; doi: https://doi.org/10.1038/nature07589.
Ferdinandusse, S., Mulders, J., Denis, S., Dacremont, G., Waterham, H. R., and Wanders, R. J. (1999) Molecular cloning and expression of human carnitine octanoyltransferase: evidence for its role in the peroxisomal β-oxidation of branched-chain fatty acids, Biochem. Biophys. Res. Commun., 263, 213–218; doi: https://doi.org/10.1006/bbrc.1999.1340.
Apryatin, S. A., Mzhel’skaya, K. V., Trusov, N. V., Balakina, A. S., Kulakova, S. N., Soto, Kh. S., Makarenko, M. A., Riger, N. A., and Tutel’yan, V. A. (2016) Comparative study of Wistar rat and C57Bl/6 mouse hyperlipidemia in vivo models, Vopr. Pitan., 85, 24–33.
Peterson, E. A., Kalikin, L. M., Steels, J. D., Estey, M. P., Trimble, W. S., and Petty, E. M. (2007) Characterization of a SEPT9 interacting protein, SEPT14, a novel testis-specific septin, Mamm. Genome, 18, 796–807; doi: https://doi.org/10.1007/s00335-007-9065-x.
Shinoda, T., Ito, H., Sudo, K., Iwamoto, I., Morishita, R., and Nagata, K. (2010) Septin 14 is involved in cortical neuronal migration via interaction with septin 4, Mol. Biol. Cell, 21, 1324–1334; doi: https://doi.org/10.1091/mbc.E09-10-0869.
Apryatin, S. A., Sidorova, Yu. S., Shipelin, V. A., Balakina, A. S., Trusov, N. V., and Mazo, V. K. (2017) Neuromotor activity, anxiety and cognitive function in the in vivo model of alimentary hyperlipidemia and obesity, Bull. Exp. Biol. Med., 163, 37–41; doi: https://doi.org/10.1007/s10517-017-3732-z.
Bengoechea-Alonso, M. T., and Ericsson, J. (2016) The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth, Cell Cycle, 15, 2753–2765; doi: https://doi.org/10.1080/15384101.2016.1220456.
Heo, H. S., Kim, E., Jeon, S. M., Kwon, E. Y., Shin, S. K., Paik, H., Hur, C. G., and Choi, M. S. (2013) A nutrigenomic framework to identify time-resolving responses of hepatic genes in diet-induced obese mice, Mol. Cells, 36, 25–38; doi: https://doi.org/10.1007/s10059-013-2336-3.
Jung, U. J., Seo, Y. R., Ryu, R., and Choi, M. S. (2016) Differences in metabolic biomarkers in the blood and gene expression profiles of peripheral blood mononuclear cells among normal weight, mildly obese and moderately obese subjects, Br. J. Nutr., 116, 1022–1032; doi: https://doi.org/10.1017/S0007114516002993.
Funding
Funding. This study was supported by the State Budget Project of the Ministry of Education and Science of Russia no. 0529-2015-0006 “Search for New Molecular Markers of Alimentary Diseases: Genomic and Post-genomic Analysis”.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval. All applicable international, national, and/or institutional guidelines for the care and use of laboratory animals were followed in this study.
Additional information
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Russian Text © The Author(s), 2019, published in Biokhimiya, 2019, Vol. 84, No. 9, pp. 1344–1358.
Rights and permissions
About this article
Cite this article
Apryatin, S.A., Trusov, N.V., Gorbachev, A.Y. et al. Comparative Whole-Transcriptome Profiling of Liver Tissue from Wistar Rats Fed with Diets Containing Different Amounts of Fat, Fructose, and Cholesterol. Biochemistry Moscow 84, 1093–1106 (2019). https://doi.org/10.1134/S0006297919090128
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297919090128