Abstract
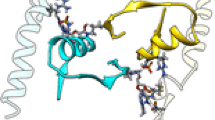
Two glycolytic enzymes, phosphoglycerate mutase (PGM) and enolase from Saccharomyces cerevisiae, have been chosen to detect complex formation and possible channeling, using molecular dynamics simulation. The enzymes were separated by 10 Å distance and placed in a water-filled box of size 173 × 173 × 173 Å. Three different orientations have been investigated. The two initial 3-phosphoglycerate substrate molecules near the active centers of the initial structure of PGM have been replaced with final product (2-phosphoglycerate) molecules, and 150 mM NaCl together with three Mg2+ ions have been added to the system to observe post-catalytic activity under near-physiological conditions. Analysis of interaction energies and conformation changes for 3 nsec simulation indicates that PGM and enolase do show binding affinity between their near active regions, which is necessary for channeling to occur. Interaction of the C-terminal residues Ala239 and Val240 of PGM (which partially “cap” the 2-phosphoglycerate) with enolase also favors the existence of channeling.
Similar content being viewed by others
Abbreviations
- MD:
-
molecular dynamics
- 2PG:
-
2-phosphoglycerate
- 3PG:
-
3-phosphoglycerate
- PGM:
-
phosphoglycerate mutase
References
Ovadi, J. (1991) J. Theor. Biol., 152, 1–22.
Agius, L., and Sherratt, H. S. A. (1997) Channeling in Intermediary Metabolism, Portland Press, London, pp. 1–11.
Clegg, J. S., and Jackson, S. A. (1989) Biochem. Biophys. Res. Commun., 160, 1409–1414.
Shearwin, K., Nanhua, C., and Masters, C. (1990) Biochem. Int., 21, 53–60.
Shearwin, K., Nanhua, C., and Masters, C. (1990) Biochem. Int., 22, 735–740.
Edwards, S. R., Braley, R., and Chaffin, W. L. (1999) FEMS Microbiol. Lett., 177, 211–216.
Motshwene, P., Brandt, W., and Lindsey, G. (2003) Biochem. J., 369, 357–362.
Batke, J. (1991) J. Theor. Biol., 152, 41–46.
Srivastava, D. K. (1991) J. Theor. Biol., 152, 93–100.
Batke, J., Nazaryan, K. B., and Karapetian, N. H. (1988) Arch. Biochem. Biophys., 264, 510–518.
Nazaryan, K., Climent, F., Simonian, S., Tompa, P., and Batke, J. (1992) Arch. Biochem. Biophys., 296, 650–653.
Ouporov, I. V., Knull, H. R., Huber, A., and Thomasson, K. A. (2001) J. Biophys., 80, 2527–2535.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Sindyalov, I. N., and Bourne, P. E. (2000) Nucleic Acids Res., 28, 235–242.
Crowhurst, G. S., Dalby, A. R., Isupov, M. N., Campbell, J. W., and Littlechild, J. A. (1999) J. Biol. Cryst., 55, 1822–1826.
Zhang, E., Brewer, J. M., Minor, W., Carreira, L. A., and Lebioda, L. (1997) Biochemistry, 36, 12526–12534.
Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S., and Karplus, M. (1983) J. Comp. Chem., 4, 187–217.
HyperChem™ Professional 7.51, Hypercube, Inc., 1115 NW 4th Street, Gainesville, Florida 32601, USA.
Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graph., 14, 33–38.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in Russian in Biokhimiya, 2006, Vol. 71, No. 4, pp. 464–470.
Originally published in Biochemistry (Moscow) On-Line Papers in Press, as Manuscript BM05-215, February 19, 2006.
Rights and permissions
About this article
Cite this article
Hakobyan, D., Nazaryan, K. Molecular dynamics simulation of interactions in glycolytic enzymes. Biochemistry (Moscow) 71, 370–375 (2006). https://doi.org/10.1134/S0006297906040043
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1134/S0006297906040043