Abstract
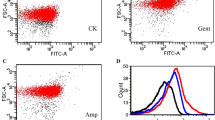
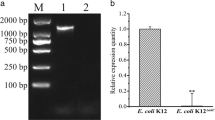
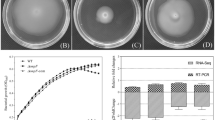
The environmental pollution caused by antibiotics is becoming more and more serious. Outer membrane proteins (OMPs) play key roles in the resistance of bacteria to antibiotics. Transcriptomic profiles of Esherichia coli treated with different types of antibiotics revealed the pathways and response mechanisms of OMP-related genes up- or down-regulated following antibiotics treatment. Among all the 4497 genes, 73 OMPs were screened. In gentamicin- or ampicillin-treated groups, 51 or 23 genes were differentially expressed, respectively. The results of protein-protein interaction networks showed that ompA, chiP, phoE, and cusC were overlapped hub genes in both groups. Considering the scores of nodes calculated by cytoHubba, OmpA was the key one among the OMPs through the calculation of Cytoscape. Finally, the knockout and overexpression mutants and complementation strain of ompA were constructed. The results of drug resistance to 4 types of antibiotics analysis confirmed the role of ompA in the resistance of E. coli to antibiotics. This work provides direct evidence for the role of outer membrane proteins, especially OmpA, in drug resistance of E. coli to different types of antibiotics. The findings in this study can lead to new strategies for improving the effectiveness of antibiotics.







Similar content being viewed by others
DATA AVAILABILITY
All data and materials used in this research are publicly available. Raw sequence data from this study have been submitted to the NCBI sequence read archive under the BioProject accession [PRJNA510855] and available at the following link: https://trace.ncbi.nlm.nih.gov/Traces/sra_sub/sub.cgi?acc=SRP173974. Other supporting data are included as additional files listed below and submitted with the manuscript.
REFERENCES
Dwyer, D.J., Belenky, P.A., Yang, J.H., MacDonald, I.C., Martell, J.D., Takahashi, N., et al., Proc. Natl. Acad. Sci. U. S. A., 2014, vol. 111, no. 20, pp. E2100–E2109. https://doi.org/10.1073/pnas.1401876111
Schlomann, B.H., Wiles, T.J., Wall, E.S., Guillemin, K., and Parthasarathy, R., Proc. Natl. Acad. Sci. U. S. A., 2019, vol. 116, no. 43, pp. 21392–21400. https://doi.org/10.1073/pnas.1907567116
Kohanski, M.A., Dwyer, D.J., Hayete, B., Lawrence, C.A., and Collins, J.J., Cell, 2007, vol. 130, no. 2, pp. 797–810. https://doi.org/10.1016/j.cell.2007.06.049
Ruddaraju, L.K., Pammi, S.V.N., Guntuku, G., Padavala, V.S., and Kolapalli, V.R.M., Asian J. Pharm. Sci., 2020, vol. 15, no. 1, pp. 42–59. https://doi.org/10.1016/j.ajps.2019.03.002
Uzoechi, S.C. and Abu-Lail, N.I., Microsc. Microanal., 2019, vol. 25, no. 1, pp. 135–150. https://doi.org/10.1017/S1431927618015696
Ormsby, M.J., Grahame, E., Burchmore, R., and Davies, R.L., J. Proteomics, 2019, vol. 199, pp. 135–147. https://doi.org/10.1016/j.jprot.2019.02.014
Horne, J.E., Brockwell, D.J., and Radford, S.E., J. Biol. Chem., 2020, vol. 295, no. 30, pp. 10340–10367. https://doi.org/10.1074/jbc.REV120.011473
Jin, M., Lu, J., Chen, Z.Y., Nguyen, S.H., Mao, L.K., Li, J.W., et al., Environ. Int., 2018, vol. 120, pp. 421–430. https://doi.org/10.1016/j.envint.2018.07.046
Xia, C.J., Li, S.F., Hou, W.Y., Fan, Z.F., Xiao, H., Lu, M.G., et al., Front. Microbiol., 2017, vol. 8, p. 2427. https://doi.org/10.3389/fmicb.2017.02427
Yu, X.J., Chen, H., Huang, C.Y., Zhu, X.Y., Wang, D.S., Liu, X.Y., et al., J. Agric. Food Chem., 2019, vol. 67, no. 19, pp. 5560–5570. https://doi.org/10.1021/acs.jafc.8b07117
Xu, M., Liu, C.L., Luo, J., Qi, Z., Yan, Z., Fu, Y., et al., BMC Genomics, 2019, vol. 20, p. 10. https://doi.org/10.1186/s12864-018-5343-0
Zheng, Y., Wang, Y., Ding, B., and Fei, Z.J., J. Virol., 2017, vol. 91, no. 11, p. e00247-17. https://doi.org/10.1128/JVI.00247-17
Li, X., Liu, X.R., Pang, X.Y., Yin, Y., Yu, H.C., Yuan, Y.X., et al., BMC Genomics, 2020, vol. 21, no. 1, p. 437. https://doi.org/10.1186/s12864-020-06850-1
Pang, X.Y., Zhao, S.J., Zhang, M., Cai, L.N., Zhang, Y.Y., and Li, X., Plant Physiol. Biochem., 2021, vol. 158, pp. 497–507. https://doi.org/10.1016/j.plaphy.2020.11.036
Amos, G.C.A., Awakawa, T., Tuttle, R.N., Letzel, A.C., Kim, M.C., Kudo, Y., et al., Proc. Natl. Acad. Sci. U. S. A., 2017, vol. 114, no. 52, pp. E11121–E11130. https://doi.org/10.1073/pnas.1714381115
Ampattu, B.J., Hagmann, L., Liang, C., Dittrich, M., Schlüter, A., Blom, J., et al., BMC Genomics., 2017, vol. 18, p. 282. https://doi.org/10.1186/s12864-017-3616-7
Sivaranjani, M., Leskinen, K., Aravindraja, C., Saavalainen, P., Pandian, S.K., Skurnik, M., et al., Front. Microbiol., 2019, vol. 10, p. 150. https://doi.org/10.3389/fmicb.2019.00150
Candar-Cakir, B., Arican, E., and Zhang, B.H., Plant Biotechnol. J., 2016, vol. 14, no. 8, pp. 1727–1746. https://doi.org/10.1111/pbi.12533
Li, X., Liu, X.R., Yin, Y., Yu, H.C., Zhang, M., Jing, H.N., et al., Food Funct., 2019, vol. 10, no. 12, pp. 8116–8128. https://doi.org/10.1039/c9fo00809h
Yang, A.M., Yu, L., Chen, Z., Zhang, S.X., Shi, J., Zhao, X.Z., et al., Viruses, 2017, vol. 9, no. 5, p. 115. https://doi.org/10.3390/v9050115
Wu, B.L., Xie, J.J., Du, Z.P., Wu, J.Y., Zhang, P.X., Xu, L.Y., et al., BioMed Res. Int., 2014, vol. 2014, p. 651954. https://doi.org/10.1155/2014/651954
Vig, S., Talwar, P., Kaur, K., Srivastava, R., Srivastava, A.K., and Datta, M., Cell Cycle, 2015, vol. 14, no. 14, pp. 2274–2284. https://doi.org/10.1080/15384101.2015.1046654
Li, Y., Cai, Z.H., Zhu, B.A., and Xu, C.S., Genes, 2018, vol. 9, no. 2, p. 92. https://doi.org/10.3390/genes9020092
Huang, H.J., Luo, B.B., Wang, B.Q., Wu, Q.W., Liang, Y.M., and He, Y., Med. Sci. Monit., 2018, vol. 24, pp. 7697–7709. https://doi.org/10.12659/MSM.912984
Ma, Z., Hu, Y.F., Liao, Z.J., Xu, J., Xu, X.H., Bechthold, A., et al., Front. Microbiol., 2020, vol. 11, p. 2074. https://doi.org/10.3389/fmicb.2020.02074
Wang, X.Y., Du, Y.L., Hua, Y., Fu, M.Q., Niu, C., Zhang, B., et al., Front. Cell. Infect. Microbiol., 2017, vol. 7, p. 410. https://doi.org/10.3389/fcimb.2017.00410
Chapman, J., Orrell-Trigg, R., Kwoon, K.Y., Truong, V.K., and Cozzolino, D., Biotechnol. Bioeng., 2021, vol 118, pp. 1511–1519. https://doi.org/10.1002/bit.27664
Gao, X.X., Yang, X.F., Li, J.H., Zhang, Y., Chen, P., and Lin, Z.L., Microb. Cell Fact., 2018, vol 17, p. 118. https://doi.org/10.1186/s12934-018-0966-z
Nie, D., Hu, Y., Chen, Z., Li, M.K., Hou, Z., Luo, X.X., et al., J. Biomed. Sci., 2020, vol. 27, no. 1, p. 26. https://doi.org/10.1186/s12929-020-0617-7
Carlson, M.L., Stacey, R.G., Young, J.W., Wason, I.S., Zhao, Z.Y., Rattray, D.G., et al., Elife, 2019, vol. 8, p. e46615. https://doi.org/10.7554/eLife.46615
Viale, A.M. and Evans, B.A., Microb. Genomics, 2020, vol. 6, no. 6, p. 000381. https://doi.org/10.1099/mgen.0.000381
Chen, Y.X., Jie, K.W., Li, B.X., Yu, H.Y., Ruan, H., Wu, J., et al., Front. Microbiol., 2020, vol. 11, p. 588952. https://doi.org/10.3389/fmicb.2020.588952
Chen, C.X., Hu, H.Z., Li, X.Q., Zheng, Z., Wang, Z.Z., Wang, X.C., et al., IEEE T. Nanobiosci., 2022, vol. 21, no. 1, pp. 37–43. https://doi.org/10.1109/TNB.2021.3105662
Lin, J., Huang, S.X., and Zhang, Q.J., Microbes Infect., 2002, vol. 4, no. 3, pp. 325–331. https://doi.org/10.1016/S1286-4579(02)01545-9
Choi, U. and Lee, C., Front. Microbiol., 2019, vol. 10, p. 953. https://doi.org/10.3389/fmicb.2019.00953
ACKNOWLEDGMENTS
We wish to thank Prof. J.A. Imlay for providing wildtype strain MG1655 for this work. We are grateful for the free online platform of Majorbio I-Sanger Cloud Platform (www.i-sanger.com). We would also like to thank English Editing by Elsevier Language Editing Services (Registration no. 331566771). This work was supported by the Natural Science.
Funding
Foundation of Guangdong Province (no. 2019A1515011685) received by Prof. Z. Lu, and Science and Technique Foundation of Henan Province (202102110296) received by Prof. X. Li.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Zhao, Q., Wu, Y., Li, B. et al. OmpA is Involved in the Early Response of Escherichia coli to Antibiotics as a Hub Outer Membrane Protein. Appl Biochem Microbiol 59, 608–621 (2023). https://doi.org/10.1134/S0003683823050204
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683823050204