Abstract
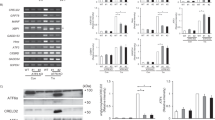
Expression of the Saccharomyces cerevisiae proteasomal genes is regulated coordinately by ScRpn4 transcription factor and its binding site known as PACE. Genomic analysis demonstrates that only Saccharomycetes yeasts carry ScRpn4-orthologous genes and PACE-like elements in the promoters of proteasomal genes. This taxonomic group includes species of biotechnological importance such as Komagataella (Pichia) pastoris and Yarrowia lipolytica. A literature search has failed to identify studies focused on ScRpn4 orthologs of these yeast species. In the present work, using the genetic background of S. cerevisiae, we showed that ScRpn4 orthologs from K. pastoris and Y. lipolytica restore the mRNA levels of proteasomal genes and confer stress tolerance in a S. cerevisiae strain with knockout of the SCRPN4 gene. In other words, these proteins are functionally similar to ScRpn4 of S. cerevisiae.
Similar content being viewed by others
Abbreviations
- a.a.:
-
amino acid
- HMF:
-
5-hydroxymethylfurfural
- A:
-
absorbance
- PCR:
-
a polymerase chain reaction
- ATP:
-
adenosine triphosphate
- dT:
-
deoxythymidine
- KpRpn4:
-
a Rpn4-like protein of Komagataella (Pichia) pastoris
- MMS:
-
a methyl methane sulfonate
- PACE:
-
proteasome-associated control element
- ScRpn4:
-
the Rpn4 gene of Saccharomyces cerevisiae
- YlRpn4:
-
a Rpn4-lijek protein of Yarrowia lipolytica
References
Glickman, M.H. and Ciechanover, A., The ubiquitinproteasome proteolytic pathway: destruction for the sake of construction, Physiol. Rev., 2002, vol. 82, pp. 373–428.
Kapranov, A.B., Kuryatova, M.V., Preobrazhenskaya, O.V., Tyutyaeva, V.V., Shtucka, R., Feldmann, H., and Karpov, V.L., Isolation and identification of PACEbinding protein Rpn4, a new transcriptional activator regulating 26S-proteasomal and other genes, Mol. Biol. (Moscow), 2001, vol. 35, pp. 356–364.
Mannhaupt, G., Schnall, R., Karpov, V., Vetter, I., and Feldmann, H., Rpn4 acts as a transcription factor by binding to pace, a nonamer box found upstream of 26A proteasomal and other genes in yeast, FEBS Lett., 1999, vol. 450, pp. 27–34.
Ju, D., Wang, L., Mao, X., and Xie, Y., Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit, Biochem. Biophys. Res. Commun., 2004, vol. 321, pp. 51–57.
Xie, Y and Varshavsky, A., Rpn4 is a ligand, substrate and transcriptional regulator of the 26S proteasome: a negative feedback circuit, Proc. Natl. Acad. Sci. USA, 2001, vol. 98, pp. 3056–3061.
Owsianik, G., Balzil, L., and Ghislain, M., Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae, Mol. Microbiol., 2002, vol. 43, pp. 1295–1308.
Teixeira, M.C., Dias, P.J., Simoes, T., and Sa-Correia, I., Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrrl, Biochem. Biophys. Res. Commun., 2008, vol. 367, pp. 249–255.
London, M., Keck, B.I., Ramos, P.C., and Dohmen, R.J., Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome, FEBS Lett., 2004, vol. 567, pp. 259–264.
Mannhaupt, G. and Feldann, H., Genomic evolution of the proteasome system among hemiascomycetous yeasts, J. Mol. Evol., 2007, vol. 65, pp. 529–540.
Ahmad, M., Hirz, M., Pichler, H., and Schwab, H., Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production, Appl. Microbiol. Biotechnol., 2014, vol. 12, pp. 5301–5317.
Goncalves, F.A., Colen, G., and Takahashi, J.A., Yarrowia lipolytica and its multiple applications in the biotechnological industry, Sci. World J., 2014, p. 476207.
Gietz, D., St.-Jean, A., Woods, R.A., and Schiestl, R.H., Improved method for high efficiency transformation of intact yeast cells, Nucleic Acids Res., 1992, vol. 20, p. 1425.
Karpov, D.S., Spasskaya, D.S., Tutyaeva, V.V., Mironov, A.S., and Karpov, V.L., Proteasome inhibition enhances resistance to DNA damage via upregulation of Rpn4-dependent DNA repair genes, FEBS Lett., 2013, vol. 587, pp. 3108–3114.
Spasskaya, D.S., Karpov, D.S., and Karpov, V.L., Escherichia coli Dam-methylase as a molecular tool for mapping binding sites of the yeast transcription factor Rpn4, Mol. Biol. (Moscow), 2011, vol. 45, pp. 591–599.
Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Smith, J.A., Seidman, J.G., and Struhl, K., Current Protocols in Molecular Biology, New York: John Wiley and Sons, 1998.
Schmitt, M.E., Brown, T.A., and Trumpower, B.L., A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae, Nucleic Acids Res., 1990, vol. 18, pp. 3091–3092.
Gasch, A.P., Moses, A.M., Chiang, D.Y., Fraser, H.B., Berardini, M., and Eisen, M.B., Conservation and evolution of cis-regulatory systems in ascomycete fungi, PLoS Biol., 2004, vol. 2, p. e398.
Vermitsky, J.-P., Earhart, K.D., Smith, W.L., Homayouni, R., Edlind, T.D., and Rogers, P.D., Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies, Mol. Microbiol., 2006, vol. 61, pp. 704–722.
Enjalbert, B., Smith, D.A., Cornell, M.J., Alam, I., Nicholls, S., Brown, A.J.P., and Quinn, J., Role of the hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans, Mol. Biol. Cell, 2006, vol. 17, pp. 1018–1032.
Ha, S.W., Ju, D., and Xie, Y., The N-terminal domain of Rpn4 serves as a portable ubiquitin-independent degron and is recognized by specific 19S RP subunits, Biochem. Biophys. Res. Commun., 2012, vol. 419, pp. 226–231.
McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y.M., Buso, N., Cowley, A.P., and Lopez, R., Analysis tool web services from the EMBL-EBI, Nucleic Acids Res., 2013, vol. 41, pp. W597–W600.
Karpov, D.S., Tutyaeva, V.V., and Karpov, V.L., Mapping of yeast Rpn4p transactivation domains, FEBS Lett., 2008, vol. 582, pp. 3459–3464.
Gonzalez-Ramos, D., van den Broek, M., van Maris, A.J., Pronk, J.T., and Daran, J.M., Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation, Biotechnol. Biofuels, 2013, vol. 6, p. 48.
Zaki, A.M., Wimalasena, T.T., and Grretham, D., Phenotypic characterization of Saccharomyces spp. for tolerance to 1-butanol, J. Ind. Microbiol. Biotechnol., 2014, vol. 41, pp. 1627–1636.
Ma, M. and Liu, Z.L., Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae, BMC Genomics, vol. 11, p. 660.
Larsson, S., Palmqvist, E., Hahn-Hagerdal, B., Tengborg, C., Stenberg, K., Zacchi, G., and Nilvebrant, N.O., The generation of inhibitors during dilute acid hydrolysis of softwood, Enz. Microb. Technol., 1999, vol. 24, pp. 151–159.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © E.N. Grineva, A.T. Leinsoo, D.S. Spasskaya, D.S. Karpov, V.L. Karpov, 2014, published in Biotekhnologiya, 2014, No. 6, pp. 8–17.
Rights and permissions
About this article
Cite this article
Grineva, E.N., Leinsoo, A.T., Spasskaya, D.S. et al. Functional analysis of Rpn4-like proteins from Komagataella (Pichia) pastoris and Yarrowia lipolytica on a genetic background of Saccharomyces cerevisiae . Appl Biochem Microbiol 51, 757–765 (2015). https://doi.org/10.1134/S0003683815070029
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683815070029