Abstract
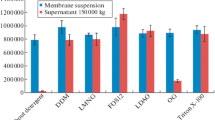
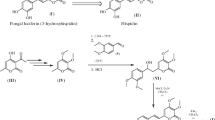
The recently described bioluminescent system from fungi has great potential for developing highly efficient tools for biomedical research. Luciferase enzyme is one of the most crucial components of this system. The luciferase from Neonothopanus nambi fungus belongs to the novel still undescribed protein family. The structure data for this protein is almost absent. A detailed study of the N. nambi luciferase properties is necessary for the improvement of analytical methods based on the fungal bioluminescent system. Here we present the positions of key amino acid residues and their effect on enzyme function described using bioinformatic and experimental approaches. These results are useful for further fungal luciferase structure determination.




Similar content being viewed by others
REFERENCES
Yan, Y., Shi, P., Song, W., and Bi, S., Chemiluminescence and bioluminescence imaging for biosensing and therapy: in vitro and in vivo perspectives, Theranostics, 2019, vol. 9, pp. 4047–4065. https://doi.org/10.7150/thno.33228
Fleiss, A. and Sarkisyan, K.S., A brief review of bioluminescent systems, Curr. Genet., 2019, vol. 65, pp. 877–882. https://doi.org/10.1007/s00294-019-00951-5
Pomper, M.G. and Gelovani, J.G., Molecular Imaging in Oncology, CRC Press, 2008.
Chew, A.L.C., Desjardin, D.E., Tan, Y.-S., et al., Bioluminescent fungi from Peninsular Malaysia—a taxonomic and phylogenetic overview, Fungal Diversity, 2015, vol. 70, pp. 149–187. https://doi.org/10.1007/s13225-014-0302-9
Shimomura, O., Bioluminescence: Chemical Principles and Methods, Singapore: World Scientific Publishing, 2006. https://doi.org/10.1142/6102
Kotlobay, A.A., Sarkisyan, K.S., Mokrushina, Y.A., et al., Genetically encodable bioluminescent system from fungi, Proc. Natl. Acad. Sci. U. S. A., 2018, vol. 115, pp. 12728–12732. https://doi.org/10.1073/pnas.1803615115
Mitiouchkina, T., Mishin, A.S., Somermeyer, L.G., et al., Plants with genetically encoded autoluminescence, Nat. Biotechnol., 2020. https://doi.org/10.1038/s41587-020-0500-9
Oliveira, A.G., Desjardin, D.E., Perry, B.A., and Stevani, C.V., Evidence that a single bioluminescent system is shared by all known bioluminescent fungal lineages, Photochem. Photobiol. Sci., 2012, vol. 11, pp. 848–852. https://doi.org/10.1039/c2pp25032b
Kaskova, Z.M., Dörr, F.A., Petushkov, V.N., et al., Mechanism and color modulation of fungal bioluminescence, Sci. Adv., 2017, p. e1602847. https://doi.org/10.1126/sciadv.1602847
Bok, J.W. and Keller, N.P., Fast and easy method for construction of plasmid vectors using modified quick-change mutagenesis, Methods Biochem. Analysis, 2012, vol. 944, pp. 163–174. https://doi.org/10.1007/978-1-62703-122-6_11
Pichia expression kit. A manual of methods for expression of recombinant proteins using pPICZ and PICZ-alpha, in Pichia pastoris Catalog, 2009. http://tools.thermofisher.com/content/sfs/manuals/pich_man.pdf.
Kwolek-Mirek, M. and Zadrag-Tecza, R., Comparison of methods used for assessing the viability and vitality of yeast cells, FEMS Yeast Res., 2014, vol. 14, pp. 1068–1079. https://doi.org/10.1111/1567-1364.12202
Funding
This work was supported by the grant from the Russian Science Foundation no. 16-14-00052P, alanine screening was performed by the President grant for leading scientific schools NSh-2605.2020.4.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any studies involving animals or human participants performed by any of the authors.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Beregovaya, K.A., Myshkina, N.M., Chepurnykh, T.V. et al. Rational Design and Mutagenesis of Fungal Luciferase from Neonothopanus nambi . Dokl Biochem Biophys 496, 14–17 (2021). https://doi.org/10.1134/S1607672921010026
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1607672921010026