Abstract
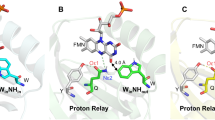
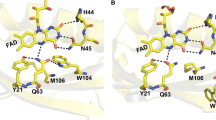
In this work we exploited time-resolved photoacoustics (PA) and molecular dynamics (MD) simulations to investigate the function of a conserved phenylalanine residue in blue sensing (BL) LOV domains. The LOV photocycle involves reversible formation of a photoproduct (LOV390) where the flavin mononucleotide (FMN) chromophore is covalently bound to a cysteine. LOV390 thermally returns to the dark adapted state (LOV447) with a lifetime τrec (s-to-h). In the LOV domain of Bacillus subtilis BsYtvA, the conserved F46 is one of the few residues undergoing a pronounced light-driven conformational change. PA and spectro-scopic data show that in the YtvA variants F46A and F46Y light-induced structural changes are much smaller than those in the wild type (wt) protein, τrec is strongly accelerated and the energy content of LOV390 is lower for F46Y. MD simulations for each variant in the LOV447 and LOV390 states revealed an overall very stable structure of the BsYtvA-LOV domain. The largest variations emerged for the conserved HB network that includes FMN, Q123 (the “flipping” glutamine of LOV domains), and the conserved N104 and N94, with strong dependence on the presence of water. The lateral chain of Q123 in wt-LOV447 can adopt three alternative conformations, and movements act in concert with F46 flexibility. In LOV390, Q123 remains instead fixed in the orientation adopted in the crystal structure. Interestingly, in F46A, Q123 is locked in a LOV447-like conformation (pseudo-dark-adapted state), in both LOV447 and LOV390. In LOV447 of F46Y the tyrosine hydroxyl group fixes a water molecule, which induces a Q123 conformation similar to wt-LOV390, i.e. a pseudo-photoproduct state. These pseudo-dark-adapted and photoproduct-like conformations of the Q123 sidechain may account for the strong acceleration of the photocycle in the two variants. Given the importance of the “flipping” glutamine in light-to-signal propagation in LOV proteins, the results presented here underscore a crucial structural and functional role of the conserved F46. MD results also indicate that F46 is not directly engaged in permeability of the FMN pocket, but is involved in solvent ordering and the formation of water bridges.
Similar content being viewed by others
References
A. Losi and W. Gärtner, Solving Blue Light Riddles: New Lessons from Flavin-binding LOV Photoreceptors, Photochem. Photobiol., 2017, 93, 141–158.
T. Fettweiss, K. Röllen, J. Granzin, O. Reiners, S. Endres, T. Drepper, D. Willbold, K.-E. Jaeger, R. Batra-Safferling and U. Krauss, Mechanistic Basis of the Fast Dark Recovery of the Short LOV Protein DsLOV from Dinoroseobacter shibae, Biochemistry, 2018, 57, 4833–4847.
S. T. Glantz, E. J. Carpenter, M. Melkonian, K. H. Gardner, E. S. Boyden, G. K.-S. Wong and B. Y. Chow, Functional and topological diversity of LOV domain photoreceptors, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E1442–E1451.
J. T. Henry and S. Crosson, Ligand-Binding PAS Domains in a Genomic, Cellular, and Structural Context, Annu. Rev. Microbiol., 2011, 65, 261–286.
S. M. Harper, L. C. Neil and K. H. Gardner, Structural Basis of a Phototropin Light Switch, Science, 2003, 301, 1541–1544.
A. Losi, E. Polverini, B. Quest and W. Gärtner, First Evidence for Phototropin-Related Blue-Light Receptors in Prokaryotes, Biophys. J., 2002, 82, 2627–2634.
A. Möglich and K. Moffat, Structural Basis for Light-dependent Signaling in the Dimeric LOV Domain of the Photosensor YtvA, J. Mol. Biol., 2007, 373, 112–126.
A. I. Nash, W.-H. Ko, S. M. Harper and K. H. Gardner, A Conserved Glutamine Plays a Central Role in LOV Domain Signal Transmission and Its Duration, Biochemistry, 2008, 47, 13842–13849.
M. Avila-Pérez, J. Vreede, Y. Tang, O. Bende, A. Losi, W. Gärtner and K. Hellingwerf, In vivo mutational analysis of YtvA from Bacillus subtilis. Mechanism of light activation of the general stress response, J. Biol. Chem., 2009, 284, 24958–24964.
A. Ganguly, W. Thiel and B. R. Crane, Glutamine Amide Flip Elicits Long Distance Allosteric Responses in the LOV Protein Vivid, J. Am. Chem. Soc., 2017, 139, 2972–2980.
A. Losi, K. H. Gardner and A. Möglich, Blue-Light Receptors for Optogenetics, Chem. Rev., 2018, 118, 10659–10709.
S. Crosson and K. Moffat, Photoexcited Structure of a Plant Photoreceptor Domain Reveals a Light-Driven Molecular Switch, Plant Cell, 2002, 14, 1067–1075.
P. L. Freddolino, K. H. Gardner and K. Schulten, Signaling mechanisms of LOV domains: New insights from molecular dynamics studies, Photochem. Photobiol. Sci., 2013, 12, 1158–1170.
M. E. Kalvaitis, L. A. Johnson, R. J. Mart, P. Rizkallah and R. K. Allemann, A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation, Biochemistry, 2019, 58, 2608–2616.
V. V. Nazarenko, A. Remeeva, A. Yudenko, K. Kovalev, A. Dubenko, I. M. Goncharov, P. Kuzmichev, A. V. Rogachev, P. Buslaev, V. Borshchevskiy, A. Mishin, G. V. Dhoke, U. Schwaneberg, M. D. Davari, K.-E. Jaeger, U. Krauss, V. Gordeliy and I. Gushchin, A thermostable flavin-based fluorescent protein from Chloroflexus aggregans : a framework for ultra-high resolution structural studies, Photochem. Photobiol. Sci., 2019, 18, 1793–1805.
J. Lokhandwala, R. I. Silverman, Y. de la Vega, H. C. Hopkins, C. W. Britton, A. Rodriguez-Iglesias, R. Bogomolni, M. Schmoll and B. D. Zoltowski, A Native Threonine Coordinates Ordered Water to Tune Light-Oxygen-Voltage (LOV) Domain Photocycle Kinetics and Osmotic Stress Signaling in Trichoderma reesei ENVOY, J. Biol. Chem., 2016, 291, 14839–14850.
A. Pudasaini, K. K. El-Arab and B. D. Zoltowski, LOV-based optogenetic devices: light-driven modules to impart photo-regulated control of cellular signaling, Front. Mol. Biosci., 2015, 2, 18.
A. Möglich and K. Moffat, Engineered photoreceptors as novel optogenetic tools, Photochem. Photobiol. Sci., 2010, 9, 1286–1300.
A. Losi and S. E. Braslavsky, The time-resolved thermodynamics of the chromophore-protein interactions in biological photosensors as derived from photothermal measurements, Phys. Chem. Chem. Phys., 2003, 5, 2739–2750.
A. Losi, T. Kottke and P. Hegemann, Recording of Blue Light-Induced Energy and Volume Changes within the Wild-Type and Mutated Phot-LOV1 Domain from Chlamydomonas reinhardtii, Biophys. J., 2004, 86, 1051–1060.
I. Kraiselburd, A. Gutt, A. Losi, W. Gärtner and E. G. Orellano, Functional Characterization of a LOV-Histidine Kinase Photoreceptor from Xanthomonas citri subsp. citri, Photochem. Photobiol., 2015, 91, 1123–1132.
R. Kaur Grewal, D. Mitra and S. Roy, Mapping networks of light–dark transition in LOV photoreceptors, Bioinformatics, 2015, 5, btv429.
R. P. Diensthuber, C. Engelhard, N. Lemke, T. Gleichmann, R. Ohlendorf, R. Bittl and A. Möglich, Biophysical, Mutational, and Functional Investigation of the Chromophore-Binding Pocket of Light-Oxygen-Voltage Photoreceptors, ACS Synth. Biol., 2014, 3, 811–819.
S.-H. Song, P. L. Freddolino, A. I. Nash, E. C. Carroll, K. Schulten, K. H. Gardner and D. S. Larsen, Modulating LOV Domain Photodynamics with a Residue Alteration outside the Chromophore Binding Site, Biochemistry, 2011, 50, 2411–2423.
K. Magerl, I. Stambolic, B. Dick, V. Balland, E. D. Getzoff, T. Ritz, K. Brettel, S. Nonell, P. J. Tonge, S. R. Meech, I. P. Clark, M. Towrie, G. M. Greetham, A. Ng, J. J. Truglio, J. B. French, S. R. Meech and P. J. Tonge, Switching from adduct formation to electron transfer in a light–oxygen–voltage domain containing the reactive cysteine, Phys. Chem. Chem. Phys., 2017, 19, 10808–10819.
S. Raffelberg, M. Mansurova, W. Gärtner and A. Losi, Modulation of the photocycle of a LOV domain photo-receptor by the hydrogen-bonding network, J. Am. Chem. Soc., 2011, 133, 5346–5356.
P. A. van den Berg, J. Widengren, M. A. Hink, R. Rigler and A. J. Visser, Fluorescence correlation spectroscopy of flavins and flavoenzymes: photochemical and photophysical aspects., Spectrochim. Acta, Part A, 2001, 57, 2135–2144.
S. Raffelberg, A. Gutt, W. Gärtner, C. Mandalari, S. Abbruzzetti, C. Viappiani and A. Losi, The amino acids surrounding the flavin 7a-methyl group determine the UVA spectral features of a LOV protein., Biol. Chem., 2013, 394, 1517–1528.
J. E. Rudzki, J. L. Goodman and K. S. Peters, Simultaneous determination of photoreaction dynamics and energetics using pulsed, time-resolved photoacoustic calorimetry, J. Am. Chem. Soc., 1985, 107, 7849–7854.
M. Gauden, S. Crosson, I. H. M. van Stokkum, R. van Gondelle, K. Moffat and J. T. M. Kennis, in Femtosecond Laser Applications in Biology, ed. S. Avrilleir and J. M. Tualle, SPIE Bellingham, WA5463, 2004, pp. 97–104.
M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindah, Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25.
A. D. MacKerell, D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, All-atom empirical potential for molecular modeling and dynamics studies of proteins., J. Phys. Chem. B, 1998, 102, 3586–3616.
P. Mark and L. Nilsson, Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K, J. Phys. Chem. A, 2001, 105, 9954–9960.
J. V. Vermaas, D. J. Hardy, J. E. Stone, E. Tajkhorshid and A. Kohlmeyer, TopoGromacs: Automated Topology Conversion from CHARMM to GROMACS within VMD, J. Chem. Inf. Model., 2016, 56, 1112–1116.
H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, The Protein Data Bank, Nucleic Acids Res., 2000, 28, 235–242.
N. Guex and M. C. Peitsch, SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling, Electrophoresis, 1997, 18, 2714–2723.
L. L. C. Schrödinger, The PyMOL Molecular Graphics System, Version 1.8, 2015, https://pymol.org.
W. Humphrey, A. Dalke and K. Schulten, VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38.
T. A. Schüttrigkeit, C. K. Kompa, M. Salomon, W. Rüdiger and M. E. Michel-Beyerle, Primary photophysics of the FMN binding LOV2 domain of the plant blue light receptor phototropin of Avena sativa, Chem. Phys., 2003, 294, 501–508.
A. Penzkofer, L. Endres, T. Schiereis and P. Hegemann, Yield of photo-adduct formation of LOV domains from Chlamydomonas reinhardtii by picosecond laser excitation, Chem. Phys., 2005, 316, 185–194.
T. Kottke, J. Heberle, D. Hehn, B. Dick and P. Hegemannt, Phot-LOV1: Photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii, Biophys. J., 2003, 84, 1192–1201.
A. Pudasaini, J. S. Shim, Y. H. Song, H. Shi, T. Kiba, D. E. Somers, T. Imaizumi and B. D. Zoltowski, Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis, eLife, 2017, 6, e21646.
A. Losi, C. Mandalari and W. Gärtner, From Plant Infectivity to Growth Patterns: The Role of Blue-Light Sensing in the Prokaryotic World, Plants, 2014, 3, 70–94.
R. Ohlendorf, R. R. Vidavski, A. Eldar, K. Moffat and A. Möglich, From dusk till dawn: One-plasmid systems for light-regulated gene expression, J. Mol. Biol., 2012, 416, 534–542.
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic supplementary information (ESI) available: Fig. S1–S10 and Tables S1–S3. See DOI: 10.1039/d0pp00082e
Rights and permissions
About this article
Cite this article
Polverini, E., Schackert, F.K. & Losi, A. Interplay among the “flipping” glutamine, a conserved phenylalanine, water and hydrogen bonds within a blue-light sensing LOV domain. Photochem Photobiol Sci 19, 892–904 (2020). https://doi.org/10.1039/d0pp00082e
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1039/d0pp00082e