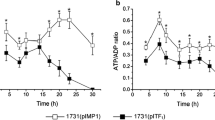
The effect of solR inactivation on the metabolism of Clostridium acetobutylicum was examined using fermentation characterization and metabolic flux analysis. The solR-inactivated strain (SolRH) of this study had a higher rate of glucose utilization and produced higher solvent concentrations (by 25%, 14%, and 81%, respectively, for butanol, acetone, and ethanol) compared to the wild type. Strain SolRH(pTAAD), carrying a plasmid-encoded copy of the bifunctional alcohol/aldehyde dehydrogenase gene (aad) used in butanol production, produced even higher concentrations of solvents (by 21%, 45%, and 62%, respectively, for butanol, acetone, and ethanol) than strain SolRH. Clarithromycin used for strain SolRH maintenance during SolRH(pTAAD) fermentations did not alter product formation; however, tetracycline used for pTAAD maintenance resulted in 90% lower solvent production.
Journal of Industrial Microbiology & Biotechnology (2001) 27, 322–328.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received 12 September 2000/ Accepted in revised form 21 July 2001
Rights and permissions
About this article
Cite this article
Harris, L., Blank, L., Desai, R. et al. Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene. J Ind Microbiol Biotech 27, 322–328 (2001). https://doi.org/10.1038/sj.jim.7000191
Issue Date:
DOI: https://doi.org/10.1038/sj.jim.7000191