Abstract
After new drugs enter the market, adverse events (AE) induced by their use must be tracked; rare AEs may not be detected during clinical trials. Some organizations have been collecting information on suspected drugs and AEs via a spontaneous reporting system to conduct post-market drug safety surveillance. These organizations use the information to detect a signal representing potential causality between drugs and AEs. The drug and AE data are often hierarchically structured. Accordingly, the tree-based scan statistic can be used as a statistical data mining method for signal detection. Most of the AE databases contain a large number of zero-count cells. Notably, not only an observational zero from the Poisson distribution, but also a true zero exists in zero-count cells. True zeros represent theoretically impossible observations or possible but unreported observations. The existing tree-based scan statistic assumes that all zeros are zero-valued observations from the Poisson distribution. Therefore, true zeros are not considered in the modeling, which can lead to bias in the inferences. In this study, we propose a tree-based scan statistic for zero-inflated count data in a hierarchical structure. According to our simulation study, in the presence of excess zeros, our proposed tree-based scan statistic provides better performance than the existing tree-based scan statistic. The two methods were illustrated using Korea Adverse Event Reporting System data from the Korea Institute of Drug Safety and Risk Management.
Similar content being viewed by others
Introduction
After new drugs enter the market, the adverse events (AE) induced by their use must be tracked because rare AEs may not be detected during clinical trials owing to short trial durations, limited sample sizes, or limited population representation. Once drugs are commercialized, they are used in different ways and by more people than those covered during clinical trials. Accordingly, drug safety must be monitored even after commercialization to identify AEs that may not have been identified previously1,2,3,4,5,6,7.
Drug and vaccine safety monitoring systems have traditionally been based on spontaneous reporting systems, such as the US Food and Drug Administration’s Adverse Event Reporting System (AERS), the US Vaccine Adverse Event Reporting System (VAERS), and VigiBase, the World Health Organization’s (WHO) global Individual Case Safety Reports database. AERS is a large database supporting the US Food and Drug Administration’s program for monitoring drug safety; VAERS helps monitor vaccine-related AEs and is maintained by the US Center for Disease Control and Prevention and the US Food and Drug Administration; and VigiBase is managed by the Uppsala Monitoring Centre (UMC) on behalf of the WHO. VigiBase receives individual case safety reports from 80 countries. In South Korea, the Korea Institute of Drug Safety and Risk Management provides information on AEs collected through the Korea Adverse Event Reporting System (KAERS) to the UMC. These spontaneous reporting systems play an important role in detecting AE signals in post-market drug safety surveillance8,9.
Disproportionality data mining methods have been used to analyze these databases to identify signs that certain drugs may be posing unrecognized safety hazards. Frequentist methods, such as the proportional reporting ratio10, relative odds ratio11, Yule’s test12, chi-squared test13, and likelihood ratio test (LRT)14, and Bayesian methods, including the Bayesian confidence propagating neural network15, multi-item gamma Poisson shrinker16, and simplified Bayes (sB) methods15,16,17,18,19 are often used to detect drugs with previously unrecognized AE16,20,21,22,23,24,25.
In pharmacovigilance data, AE information uses adverse reaction terms, which have a hierarchical structure. For example, as shown in Fig. 1, the WHO Adverse Reaction Terminology (WHO-ART) developed for the WHO drug monitoring program has a four-level hierarchical structure. (https://www.who-umc.org) Owing to this type of structure, it is difficult to determine the level of AE definition that should be used during data mining. To solve the problem, tree-based scan statistics, which find signals at each level of AEs in the form of a hierarchical tree, have been proposed by Kulldorff et al.26 and have been recently used by some researchers to detect AE signals27,28,29. The tree-based scan statistic is distinct from most disproportionality methods; it is based on scan statistical theory and uses a hierarchical diagnosis tree to simultaneously assess risk at any level of granularity, adjusting for a multiple testing problem in several overlapping evaluated groups7,26,30.
Most of these AE databases have large numbers of zero-count cells. For example, AERS data from 2006 to 2011 show that the percentage of zero-count cells by the drug ranges from 50 to 99.99%31. However, based on KAERS data from 2012 to 2016, the percentage of zero-count cells by the drug ranges from 75 to 100%. Zero-count cells may contain not only zero-valued observations from the Poisson distribution, but also true zeros, which represent theoretically impossible observations or possible but unreported observations. Data with a large number of zeros cannot be assumed to have a Poisson distribution as some zeros are true zeros. The distribution of such data is typically more dispersed than the Poisson distribution, resulting in equality between the variance and the mean of the distribution. To solve this problem, the zero-inflated Poisson (ZIP) model proposed by Lambert32 can be used. Huang et al.31,33 proposed a zero-inflated Poisson model based likelihood ratio test (ZIP-LRT) method as an extended version of LRT, a frequentist data mining method. Further, Hu et al.24 developed the zero-inflated Poisson simplified Bayes method and the zero-inflated Poisson Dirichlet process method, which are Bayesian data mining methods.
The existing tree-based scan statistic assumes all zero values are zero-valued observations from the Poisson distribution. As a result, true zeros are not considered in the modeling, which can lead to bias in the inferences. Therefore, in this study, we proposed a new tree-based scan statistic using the ZIP model for data with excess zeros in a hierarchical structure.
In section “A tree-based scan statistic”, we introduce the existing tree-based scan statistic. In section “A tree-based scan statistic for zero inflated count data”, we propose a tree-based scan statistic for zero-inflated count data. In section “Simulation study”, a simulation study to evaluate the performance of the proposed method is presented. In section “Real data”, the two methods are compared through a real data example. Finally, in section “Conclusion and discussion”, we summarize the results and conclude with our recommendations.
Hierarchical diagnosis tree
The tree-based scan statistic uses hierarchical classification systems to represent clinical concepts, such as drugs, procedures, or diagnoses30. To code adverse drug reactions in postmarket drug surveillance, medical terminologies, such as Medical Dictionary for Regulatory Activities (MedDRA) and WHO-ART, are used. In the KEARS data, WHO-ART is used to code the AEs.
WHO-ART is the terminology for coding clinical information related to pharmacotherapy and is commonly used for coding the AEs. When new drugs and new symptoms create new terms that incorporate them, the structure of the terms is updated to include the newly integrated terms while retaining their previous relationships and the existing structure of terms. WHO-ART has a four-level hierarchical structure, which consists of System Organ Class (SOC), High Level Terms (HLT), Preferred Terms (PT), and Included Terms (IT). The highest level, the SOC, corresponds to body systems and organs, which contain grouping terms. The HLT is used to group related or similar PTs, but all PTs are not grouped into the HLT. The PTs are principal terms used to describe AEs and the ITs are synonyms of the PTs, which help in the search for the PTs. An example of the WHO-ART is shown in Table 1.
A tree-based scan statistic
Review of a tree-based scan statistic
The tree-based scan statistic is a statistical data mining method that has been used for signal detection in a hierarchically structured data, such as a classification system for coding AEs. This statistic searches signals at any level of AE definitions, called leaves. Each leaf contains information on the total number of patients with a specific AE and the number of patients with a specific AE from a certain drug. Mutually-related leaves are grouped into a higher level, called a node. Of note, a cut defines a branch of the tree where a node or a leaf may have more events than expected.
The tree-based scan statistic method considers all possible cuts. For each cut, the total number of AEs from all drugs and a certain drug are respectively calculated for the leaves within that cut. The test statistic is generated by a likelihood function in which risk is estimated separately for the leaves defined by the cut and those outside of the cut26,34,35.
Let \({c}_{i}\) be the observed number of patients with ith AE potentially caused by a certain drug in leaf \(i\) and \({n}_{i}\) be the total observed number of patients with \(i\) th AE in leaf \(i\). For a rare disease, with covariates ignored, \({c}_{i}\) is approximately Poisson distributed with mean \({n}_{i}{\lambda }_{i}\), where \({\lambda }_{i}\) is the probability that \(i\) th AE is caused by a certain drug. For all leaves on the tree, let \(C={\sum }_{i=1}^{I}{c}_{i}\) and \(N={\sum }_{i=1}^{I}{n}_{i}\) where I is the number of all leaves in the tree. For each cut G, a leaf or a group of related leaves, let \({c}_{G}={\sum }_{i\in G}{c}_{i}\) and \({n}_{G}={\sum }_{i\in G}{n}_{i}\). R is the rest of the leaves except those included in G. The following null hypothesis \({H}_{0}: {\lambda }_{G}={\lambda }_{0}\) and the alternative hypothesis \({H}_{a}:{\lambda }_{G}>{\lambda }_{R}\) are considered. The null hypothesis suggests that the probability that AEs in a cut G due to a certain drug are not lower or higher than that of all AEs. The alternative hypothesis is that at least one cut is defined by a set G such that \({\lambda }_{G}>{\lambda }_{R}\), where R is a group of the remaining leaves.
Of note, the analysis is only concerned with C, as the total number of AEs represented by the tree is not of interest. In fact, only the relative distribution between the different AEs is relevant. The likelihood can then be expressed as \(L\left(\lambda ,{\varvec{c}}\right)=\prod_{i}{\left(\frac{{n}_{i}{\lambda }_{i}}{{\sum }_{i}{n}_{i}{\lambda }_{i}}\right)}^{{c}_{i}}\) using a multinomial distribution. As a maximum likelihood estimator (MLE) of \({\lambda }_{G}/{\lambda }_{R}\) is \(\frac{{c}_{G}/{n}_{G}}{(C-{c}_{G})/(N-{n}_{G})}\) given G, a likelihood ratio test statistic is \(T= \frac{\underset{G, {\lambda }_{G}>{\lambda }_{R}}{\mathrm{max}}L\left(\lambda , {\varvec{c}}\right)}{\underset{ {\lambda }_{G}={\lambda }_{R}}{\mathrm{max}}L\left(\lambda , {\varvec{c}}\right)}={\left(\frac{N}{C}\right)}^{C}\underset{G}{\mathrm{max}}{\left(\frac{{c}_{G}}{{n}_{G}}\right)}^{{c}_{G}}{\left(\frac{C-{c}_{G}}{N-{n}_{G}}\right)}^{C-{c}_{G}}\) when \(\frac{{c}_{G}}{{n}_{G}}>\frac{C-{c}_{G}}{N-{n}_{G}}\); otherwise, the statistic is 1. The log-likelihood ratio-based test statistic is given by
where I() is the indicator function26.
Hypothesis testing
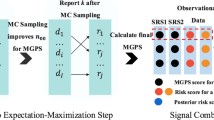
To calculate the test statistic T, the likelihood of each possible cut was determined. The cut, which is maximizing the likelihood ratio value, is defined as the most likely cut; the likelihood ratio value is defined as the test statistic T. As the null distribution of the test statistic is unknown, it is produced using the Monte Carlo simulation36. Given the total number of patients with AEs from a certain drug, a large number of random data sets was created under the null hypothesis, and the test statistics for each random data set and the real data were calculated. The obtained test statistics for random datasets were compared to the test statistic for the real data. The P-value was calculated using the equation: rank/(1 + B), where rank is the relative position of the test statistic for the real data among the test statistics for the random data sets and B is the number of Monte Carlo replications.
A tree-based scan statistic for zero-inflated count data
In the presence of excess zero, the Poisson model tends to underestimate the observed dispersion. In this case, the ZIP model can be employed as one of the approaches to resolve the problem as this model is more flexible than the Poisson model. If the number of ith AE with a certain drug \({C}_{i}\) follows the ZIP model, with the probability p of a true zero and the average number of events \({n}_{i}{\lambda }_{i}\), \({C}_{i} \sim \mathrm{ZIP}(p, {n}_{i}{\lambda }_{i})\), the mean and variance can be expressed as \(E({C}_{i}\left|p, {n}_{i}{\lambda }_{i}\right)=(1-p){n}_{i}{\lambda }_{i}\) and \(V({C}_{i}\left|p, {n}_{i}{\lambda }_{i}\right)=(1-p){n}_{i}{\lambda }_{i}(1+p{n}_{i}{\lambda }_{i})\). It can also be expressed as \(V({C}_{i}\left|p, {n}_{i}{\lambda }_{i}\right)=E({C}_{i}\left|p, {n}_{i}{\lambda }_{i}\right)(1+p{n}_{i}{\lambda }_{i})\); thus, \(V({C}_{i}\left|p, {n}_{i}{\lambda }_{i}\right)>E({C}_{i}\left|p, {n}_{i}{\lambda }_{i}\right)\) when p > 0.
As the ZIP model has an additional parameter relative to the tree-based scan statistic, its mean is smaller than that of the Poisson model. Thus, the ZIP model correctly calculates a reduced number of ith AEs with a certain drug due to the presence of true zeros.
Given the parameters \(p\) and \({n}_{i}{\lambda }_{i}\), the probability of \({C}_{i}={c}_{i}\) is described as follows:
For the tree-based ZIP scan statistic, the hypotheses of interest are the same as those in section “Review of a tree-based scan statistic”. The zeros are assumed to be known, whether or not they are true zeros, as it is difficult to find a closed form of MLE when the nature of each zero is unknown. As tree-based scan statistics are based on scan statistic theory, the methodology of Cançado et al.37, who proposed a spatial scan statistical method for zero-inflated Poisson processes, was employed.
We consider a vector \(\delta =({\delta }_{1},\dots , {\delta }_{I})\) where \({\delta }_{i}=1\) for a true zero in leaf \(i\) and \({\delta }_{i}=0\) for an observational zero in leaf \(i\). \({\delta }_{i}\) s are Bernoulli random variables with the probability p of a true zero. Given a set of observations \(\delta =({\delta }_{1},\dots ,{\delta }_{I})\) that are bivariate data such that \(\left({C}_{i}, {\delta }_{i}\right)\), \(i=1,\dots ,I\), the likelihood function for set G can be expressed as
When \({\delta }_{i}\) s are known, the MLEs under the null hypothesis are \({\widehat{\lambda }}_{0}=\frac{{\sum }_{i=i}^{I}{c}_{i}\left(1-{d}_{i}\right)}{{\sum }_{i=i}^{I}{n}_{i}\left(1-{d}_{i}\right)}\mathrm{ and }{\widehat{p}}_{0}=\frac{{\sum }_{i=i}^{I}{d}_{i}}{I}\). However, under the alternative hypothesis, the MLEs are \({\widehat{\lambda }}_{G}=\frac{{\sum }_{i\in G}{c}_{i}\left(1-{d}_{i}\right)}{{\sum }_{i\in G}{n}_{i}\left(1-{d}_{i}\right)}, {\widehat{\lambda }}_{R}=\frac{{\sum }_{i\notin G}{c}_{i}\left(1-{d}_{i}\right)}{{\sum }_{i\notin G}{n}_{i}\left(1-{d}_{i}\right)},\mathrm{ and }\widehat{p}=\frac{{\sum }_{i=i}^{I}{d}_{i}}{I}\).
When \({\delta }_{i}\) s are unknown, an expectation–maximization (EM) algorithm is used to find the MLEs of \({\lambda }_{0},{\lambda }_{G}, {\lambda }_{R}, {p}_{0}\) and \(p\). In the expectation step (E-step), the expected value of \({\delta }_{i}\), given \({C}_{i}\), is calculated using the following formula:
Under \({H}_{a}\), \({\widehat{\lambda }}_{0}\) is considered \({\widehat{\lambda }}_{G}\) and \({\widehat{\lambda }}_{R}\) in each cut G and the remaining leaves R, respectively.
In the maximization step (M-step), the MLEs of \({\lambda }_{0},{\lambda }_{G}, {\lambda }_{R}, {p}_{0}\), and \(p\) are updated via the equations with \({d}_{i}\) replaced by \({{\widehat{\delta }}_{i}}^{\left(m\right)}\) when \({\delta }_{i}\) s are known. Until the maximum likelihood estimates for each possible cut G converge, the above E- and M-steps are performed repeatedly. To perform a faster calculation, we used the ‘zeroinfl’ function in the R package “pscl”38. For the possible candidate cuts, this process should be conducted and the most likely cut should be determined.
The likelihood ratio for cut G can be expressed as
Thereafter, the maximum likelihood ratio is defined as the test statistic, \(T=\underset{G}{\mathrm{max}}L{R}_{G}.\)
As it is impossible to know the null distribution of the likelihood ratio test statistic T, Monte Carlo hypothesis testing was conducted to assess statistical significance37.
Simulation study
Data generating process and performance assessment measures
We conducted a simulation study to assess the performance of the proposed tree-based scan statistic for zero-inflated count data (TreeScan-ZIP) and the existing tree-based scan statistic (TreeScan-Poisson). For the simulation study, datasets with the hierarchical structure where AEs can be expressed in terms of WHO-ART SOCs and PTs were generated. Only 105 of the 1292 AEs in the PT terms were considered to reduce computation time. Different artificial true signals and true zeros were generated using a tree with 105 leaves and 9 nodes. The total numbers of patients with each AE varied from 10 to 4670. The total number of patients in all leaves of the tree was 19,920 and the total number of patients with AEs from a certain drug was 640.
First, true zeros \(({\delta }_{i}=1)\) were randomly allocated using the Bernoulli distribution with the probability p, where p is the percentage of the true zero leaves. Thereafter, for each iteration, the total number of patients with AEs from a certain drug, that is \(C={\sum }_{i=1}^{I}{c}_{i}\), was randomly assigned to the leaves on the tree as multinomial, with probabilities proportional to the relative risk. The relative risk of ith leaf was computed as \(\frac{{c}_{i}/{n}_{i}}{C/N}, i=1,\dots ,I.\) For true zero leaves, \(\left({\delta }_{i}=1\right), {c}_{i}=0\). If the ith leaf was not a true zero, the dataset was generated using a multinomial distribution. Under H0, the vector \({\varvec{C}}=({c}_{1},\dots , {c}_{I})\) follows a multinomial distribution with parameters \({\varvec{C}}\) and \({\varvec{p}}\), where \({\varvec{p}}=\left(\frac{{n}_{1}}{N}, \dots ,\frac{{n}_{I}}{N}\right)\). Under \({H}_{a}\), \({\varvec{p}}=\left(\frac{r{r}_{1}\frac{{n}_{1}}{N}}{{\sum }_{i=1}^{I}r{r}_{i}\frac{{n}_{i}}{N}}, \dots , \frac{r{r}_{I}\frac{{n}_{I}}{N}}{{\sum }_{i=1}^{I}r{r}_{i}\frac{{n}_{i}}{N}}\right)\), where \(r{r}_{1},\dots ,r{r}_{I}\) are the relative risks of all types of AEs. The relative risk of the randomly selected true signal leaves ranged from 3, 4, and 2 to 6; however, for the other leaves, except the true zero leaves, the relative risk was equal to 1.
Based on the total number of cases, C = 640, we considered 0, 10, 30, 50, and 70 for the number of true zero leaves, and 1%, 3%, 5%, and 10% for the true signal leaves with the relative risk (RR). All possible combinations were simulated.
To evaluate the performance of the two methods, we computed type I error, power, sensitivity, and positive predicted value (PPV). First, the critical value T* was obtained from 10,000 random datasets under H0 by the Monte Carlo replications for each scenario according to the number of true zeros (0, 10, 30, 50, 70). Thereafter, B random datasets were generated under \({H}_{0}\) and \({H}_{a}\) to calculate type I error, power, sensitivity, and PPV. For each of the B random datasets, test statistic \({T}_{k}, k=1,\dots ,B,\) was calculated using both methods.
Thereafter, type I error and power were estimated using
Sensitivity and PPV for each random datasets are expressed as
Overall sensitivity and PPV were calculated as the average of sensitivity and PPV over \({B}^{^{\prime}}\) random datasets, where \({B}^{^{\prime}}={\sum }_{k=1}^{B}I({T}_{k}>{T}^{*})\).
Results
The results obtained using the simulated data are presented in Table 2. The type I errors for the TreeScan-Poisson and TreeScan-ZIP methods were close to 0.05, except when the data had a Poisson distribution. The type I error of the TreeScan-Poisson method was above the nominal significance level of 0.05, while the type I error of the TreeScan-ZIP method tended to be less than 0.05.
When the data did not include true zeros (i.e., the data were generated from the Poisson distribution), the TreeScan-Poisson and TreeScan-ZIP methods produced similar power, sensitivity, and PPV estimates.
The TreeScan-ZIP method was identified to produce higher power and sensitivity estimates than the TreeScan-Poisson method when the number of true zeros was greater than or equal to 10. In the presence of zero inflation, when the number of true signals was greater than or equal to 5 and the RR was high, the PPV of the TreeScan-Poisson method was 1.0. The TreeScan-Poisson method could detect highly significant cuts, resulting in a small number of detected signals, which indicated high PPV and low sensitivity.
The TreeScan-ZIP method performed better than the TreeScan-Poisson in every dataset with true zero. The estimated power was almost 1.0 and the PPV was greater than 0.98 when the number of true zeros was greater than or equal to 10 and the number of true signals was greater than or equal to 5. The TreeScan-ZIP method was more sensitive than the TreeScan-Poisson method. The sensitivity and PPV of the TreeScan-ZIP method became higher with higher RR. When two true signals existed, both methods had a relatively low power; however, the power of the TreeScan-ZIP method increased as the number of true zeros and RR increased.
The simulation study showed that in the presence of zero inflation, the TreeScan-ZIP method performed better than the TreeScan-Poisson method.
Real data
Korea adverse event reporting system data
KAERS is a spontaneous AE reporting system maintained by the Korea Institute of Drug Safety and Risk Management (https://www.drugsafe.or.kr). Consumers, Healthcare Professionals, Regional Pharmacovigilance Centers (RPVCs), and pharmaceutical companies can report suspected drug information and AE information using the KAERS. RPVCs evaluate causality between the suspected drug and AE and report them to KIDS. The information is then stored in the KAERS as an individual case safety report (ICSR), which contains information on suspected drug, AE, causal relationship, and demographic. The ICSRs are periodically summited to the WHO-UMC. Further, safety information obtained from KAERS data and signal analysis is periodically reported to the Ministry of Food and Drug Safety.
For the real data analysis, data cleansing was performed. Because a certain drug and AE information can be reported multiple times depending on the dose and time of administration, if the same drug and AE were reported twice or more, only the first report was used. In the causality, only drug–AE pairs that received ratings of possible or above were included in this study. There are 6 levels of causality: certain, probable, possible, unlikely, conditional, and unassessable39,40. In KAERS database, AEs are coded by the WHO-ART. As more than half of the reports included information down to the PT level, and HLT may not exist, this study used two levels of hierarchy, SOC and PT, with the exception of the HLT and IT level.
Data obtained between 2012 and 2016 from KAERS were used. During this period, 716,584 people reported experiencing AEs. There were 1.8 million drug reports on 1981 types of drugs and 1.1 million AE reports on 4078 types of AEs. Further, a total of 2.4 million unique drug-AE pairs were found. When removing pairs that had beneath the ‘possible’ threshold, the final dataset analyzed in this study included 1,077,060 drug-AE pairs representing 1292 types of AEs in PTs. Further, 1981 types of drugs were identified in 557,390 reports.
Paclitaxel and docetaxel
The two proposed methods were applied to detect the AE signals to the drug–AE pairs data from KAERS. Paclitaxel and docetaxel, which have the highest sales among all anticancer drugs in the world, were selected41. Of note, these are representatives of the new class of taxane drugs, which have emerged as a fundamental treatment for breast cancer. Paclitaxel and docetaxel have similar main structures and mechanisms of action42. Paclitaxel is used to treat a number of cancer types, including Kaposi sarcoma, breast cancer, ovarian cancer, lung cancer, cervical cancer, and pancreatic cancer (https://www.ashp.org/). Docetaxel is also used as to treat several cancer types, including breast cancer, non-small cell lung cancer, prostate cancer, head and neck cancer, and stomach cancer (https://www.cancer.gov/). The most frequently reported AEs related to taxene from MICROMEDEX® include cardiovascular effects, dermatologic effects, endocrine/metabolic effects gastrointestinal effects, hematologic effects, hepatic effects, immunologic effects, musculoskeletal effects, neurologic effects, ophthalmic effects, otic effects, renal effects, respiratory effects, and others (https://www.who.int/).
Results
Paclitaxel
Nine signals were identified by the TreeScan-Poisson method and 30 signals were detected by the TreeScan-ZIP method (Table 3). The nine signals detected by the TreeScan-Poisson method were also detected by the TreeScan-ZIP method. The AEs corresponding to the signals found by both methods were related to the following SOCs: central & peripheral nervous system disorders (0410), respiratory system disorders (1100), white cell and reticuloendothelial system disorders (1220), and body as a whole—general disorders (1810). Further, their PTs were paresthesia (0410.0137), neuropathy peripheral (0410.1313), dyspnea (1100. 0514), granulocytopenia (1220.0572), leucopenia (1220.0908), chest pain (1810.0718), and temperature change sensations (1810.1705). The TreeScan-ZIP method detected signals related to 10 SOC terms. The nine signals detected by the two methods were included in the known AEs. However, some signals detected by TreeScan-ZIP alone were included in the known AEs.
Docetaxel
The TreeScan-Poisson and the TreeScan-ZIP methods identified 9 and 56 signals, respectively (Table 4). All signals detected by the TreeScan-Poisson method were also detected by the TreeScan-ZIP method. The AEs corresponding to the signals found by both methods were related to the following SOCs: skin and appendages disorders (0100), musculo-skeletal system disorders (0200), central & peripheral nervous system disorders (0410), red blood cell disorders (1210), white cell and reticulo-endothelial system (RES) disorders (1220). Their PTs were alopecia (0100.0002), nail disorder (0100.0020), myalgia (0200.0073), sensory disturbance (0410.0148), anemia (1210.0544), and granulocytopenia (1220.0572). The TreeScan-ZIP method detected signals related to 18 SOC terms. All signals detected by the two methods were included in the known AEs. A few signals that were not detected by TreeScan, but were detected by TreeScan-ZIP, were included in known AEs, such as vision disorders, gastro-intestinal system disorders, liver and biliary system disorders, urinary system disorders, etc.
Conclusion and discussion
This study sought to reveal how the tree-based scan statistic developed by Kulldorff et al.26 can be extended for the zero-inflated count data. To consider a large number of zero cells, we proposed the TreeScan-ZIP method, which integrates a zero-inflated Poisson model into the TreeScan-Poisson method. Herein, a simulation study was conducted with different settings for the relative risk and the number of true zero leaves and true signal leaves. Based on the findings of the simulation study, the TreeScan-ZIP method performed better than the TreeScan-Poisson method in terms of power, sensitivity, and PPV, especially when the proportion of true zeros was high. The real data examples also supported the simulation results. The TreeScan-Poisson method may have missed many signals that were detected by the TreeScan-ZIP method in datasets with a large number of true zeros. If the TreeScan-ZIP method detects too many false positive signals, it may increase confusion in further investigation and utilize unnecessary energy. However, even the known AEs were not detected by the TreeScan-Poisson method. Although we do not know whether all signals detected by the TreeScan-ZIP method were true, it is safer to over-detect than to miss any signal in drug safety surveillance.
The data used were extracted from spontaneous reporting systems, which is a limitation. As spontaneous reporting systems are based on self-reporting by people, such as consumers and healthcare professionals, underreporting or overreporting of AEs may easily occur. For example, only the number of cases reported can be known. Thus, whether the same AE occurred multiple times in the same person cannot be known. Cases of overreporting may thus lead to bias in the analysis.
In this study, the TreeScan-ZIP method and TreeScan-Poisson method identified signals of AEs for a particular drug, and could identify drugs that are more frequently reported to be related to a particular AE. Cuts were made either above or below nodes in this study; however, more elaborate cuts, such as the combinational cuts proposed by Kulldorff et al.7 can also be made. In this study, we used a two-level structure; however, structures with more than two levels or other spontaneous reporting system data with more delicate levels can be employed. Further studies could use a zero-inflated double Poisson or zero-inflated negative binomial model to accommodate large numbers of true zeros and overdispersion43. When a priory level of AE definition cannot be determined in the tree structure and the data have a large number of zeros, the proposed tree-based scan statistic can serve as a very useful method for detecting signals in the post-market drug safety surveillance.
Data availability
The KARES database is provided via the Korea Institute of Drug Safety and Risk management webpage. (https://open.drugsafe.or.kr/original/invitation.jsp) upon request.
References
Baciu, A., Stratton, K., Burke, S.P. Committee on the Assessment of the US Drug Safety System. The Future of Drug Safety: Promoting and Protecting the Health of the Public (2006).
Platt, R. et al. The new Sentinel network—improving the evidence of medical-product safety. N. Engl. J. Med. 361(7), 645–647 (2009).
Avorn, J. & Schneeweiss, S. Managing drug-risk information—what to do with all those new numbers. N. Engl. J. Med. 361(7), 647–649 (2009).
Davis, R. L. et al. Active surveillance of vaccine safety: A system to detect early signs of adverse events. Epidemiology 16(3), 336–341 (2005).
Platt, R. et al. Multicenter epidemiologic and health services research on therapeutics in the HMO research network center for education and research on Therapeutics. Pharmacoepidemiol. Drug Saf. 10(5), 373–377 (2001).
Yih, W. K. et al. Active surveillance for adverse events: The experience of the vaccine safety Datalink project. Pediatrics 127(Supplement 1), S54–S64 (2011).
Kulldorff, M. et al. Drug safety data mining with a tree-based scan statistic. Pharmacoepidemiol. Drug Saf. 22(5), 517–523. https://doi.org/10.1002/pds.3423 (2013).
Singleton, J. A. et al. An overview of the vaccine adverse event reporting system (VAERS) as a surveillance system. Vaccine. 17(22), 2908–2917 (1999).
Lindquist, M. VigiBase, the WHO global ICSR database system: Basic facts. Drug Inf. J. 42(5), 409–419 (2008).
Evans, S. J., Waller, P. C. & Davis, S. Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol. Drug Saf. 10(6), 483–486. https://doi.org/10.1002/pds.677 (2001).
Rothman, K. J., Lanes, S. & Sacks, S. T. The reporting odds ratio and its advantages over the proportional reporting ratio. Pharmacoepidemiol. Drug Saf. 13(8), 519–523. https://doi.org/10.1002/pds.1001 (2004).
Udny, Y., Kendall, M.G., et al. An introduction to the theory of statistics. An introduction to the theory of statistics (14th ed). (1950).
Greenwood, P. E. & Nikulin, M. S. A guide to chi-squared testing (Wiley, New York, 1996).
Huang, L., Zalkikar, J. & Tiwari, R. C. A likelihood ratio test based method for signal detection with application to FDA’s drug safety data. J. Am. Stat. Assoc. 106(496), 1230–1241 (2011).
Bate, A. et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur. J. Clin. Pharmacol. 54(4), 315–321. https://doi.org/10.1007/s002280050466 (1998).
DuMouchel, W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. Am. Stat. 53(3), 177–190. https://doi.org/10.1080/00031305.1999.10474456 (1999).
Huang, L., Zalkikar, J. & Tiwari, R. C. Likelihood ratio test-based method for signal detection in drug classes using FDA’s AERS database. J. Biopharm. Stat. 23(1), 178–200. https://doi.org/10.1080/10543406.2013.736810 (2013).
Noren, G. N., Bate, A., Orre, R. & Edwards, I. R. Extending the methods used to screen the WHO drug safety database towards analysis of complex associations and improved accuracy for rare events. Stat. Med. 25(21), 3740–3757. https://doi.org/10.1002/sim.2473 (2006).
DuMouchel, W., Pregibon, D. Empirical bayes screening for multi-item associations. Proceedings of the seventh ACM SIGKDD international conference on Knowledge discovery and data mining; San Francisco, California: Association for Computing Machinery 67–76 (2001).
Cesana, M. et al. Bayesian data mining techniques: The evidence provided by signals detected in single-company spontaneous reports databases. Drug Inf. J. 41(1), 11–21 (2007).
O’Neill, R. T. & Szarfman, A. Some US food and drug administration perspectives on data mining for pediatric safety assessment. Curr. Ther. Res. 62(9), 650–663 (2001).
Szarfman, A., Machado, S. G. & O’Neill, R. T. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA’s spontaneous reports database. Drug Saf. 25(6), 381–392 (2002).
Poluzzi, E., Raschi, E., Moretti, U. & De Ponti, F. Drug-induced torsades de pointes: data mining of the public version of the FDA Adverse Event Reporting System ( AERS ). Pharmacoepidemiol. Drug Saf. 18(6), 512–518 (2009).
Hu, N., Huang, L. & Tiwari, R. C. Signal detection in FDA AERS database using Dirichlet process. Stat. Med. 34(19), 2725–2742. https://doi.org/10.1002/sim.6510 (2015).
Wilson, A. M., Thabane, L. & Holbrook, A. Application of data mining techniques in pharmacovigilance. Br. J. Clin. Pharmacol. 57(2), 127–134 (2004).
Kulldorff, M., Fang, Z. & Walsh, S. J. A tree-based scan statistic for database disease surveillance. Biometrics 59(2), 323–331 (2003).
Lee, H., Kim, J. H., Choe, Y. J. & Shin, J.-Y. Safety surveillance of pneumococcal vaccine using three algorithms: Disproportionality methods, empirical bayes geometric mean, and tree-based scan statistic. Vaccines. 8(2), 242 (2020).
Kim, J. H., Lee, H. & Shin, J.-Y. Bacillus Calmette-Guérin (BCG) vaccine safety surveillance in the Korea adverse event reporting system using the tree-based scan statistic and conventional disproportionality-based algorithms. Vaccine. 38(21), 3702–3710 (2020).
Kim, S. et al. Data-mining for detecting signals of adverse drug reactions of fluoxetine using the Korea adverse event reporting system (KAERS) database. Psychiatr. Res. 256, 237–42. https://doi.org/10.1016/j.psychres.2017.06.038 (2017).
Wang, S. V. et al. Data mining for adverse drug events with a propensity score-matched tree-based scan statistic. Epidemiology 29(6), 895–903. https://doi.org/10.1097/ede.0000000000000907 (2018).
Huang, L., Zalkikar, J. & Tiwari, R. Likelihood ratio based tests for longitudinal drug safety data. Stat. Med. 33(14), 2408–2424 (2014).
Lambert, D. Zero-inflated Poisson regression, with an application to defects in manufacturing. Technometrics 34(1), 1–14 (1992).
Huang, L., Guo, T., Zalkikar, J. N. & Tiwari, R. C. A review of statistical methods for safety surveillance. Ther. Innov. Regul. Sci. 48(1), 98–108. https://doi.org/10.1177/2168479013514236 (2014).
Loader, C. R. Large-deviation approximations to the distribution of scan statistics. Adv. Appl. Probab. 23(4), 751–771 (1991).
Kulldorff, M. A spatial scan statistic. Commun. Stat.-Theory Methods. 26(6), 1481–1496 (1997).
Dwass, M. Modified randomization tests for nonparametric hypotheses. Ann. Math. Stat. 181–7 (1957).
Cançado, A. L., Da-Silva, C. Q. & Da Silva, M. F. A spatial scan statistic for zero-inflated Poisson process. Environ. Ecol. Stat. 21(4), 627–50 (2014).
Jackman, S. Package ‘pscl’. Technical Report. (Stanford, CA: Political Science Computational Laboratory, Stanford University, 2006).
Edwards, I. R. & Biriell, C. Harmonisation in pharmacovigilance. Drug Saf. 10(2), 93–102 (1994).
Edwards, I. R. & Aronson, J. K. Adverse drug reactions: Definitions, diagnosis, and management. The Lancet. 356(9237), 1255–1259 (2000).
Wang, L., Du, G.-H. Paclitaxel. Natural Small Molecule Drugs from Plants. 537–43 (2018).
Verweij, J., Clavel, M. & Chevalier, B. Paclitaxel (Taxol) and docetaxel (Taxotere): Not simply two of a kind. Ann Oncol. 5(6), 495–505. https://doi.org/10.1093/oxfordjournals.annonc.a058903 (1994).
de Lima, M.S., Duczmal, L.H., Neto, J.C., Pinto, L.P. Spatial scan statistics for models with overdispersion and inflated zeros. Stat. Sinica. 225–41 (2015).
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019R1F1A1057182).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by G.P. The first draft of the manuscript was written by G.P. and I.J. commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, G., Jung, I. A tree-based scan statistic for zero-inflated count data in post-market drug safety surveillance. Sci Rep 12, 16299 (2022). https://doi.org/10.1038/s41598-022-19998-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19998-5
- Springer Nature Limited