Abstract
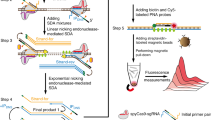
Rapid detection of nucleic acids is integral to applications in clinical diagnostics and biotechnology. We have recently established a CRISPR-based diagnostic platform that combines nucleic acid pre-amplification with CRISPR–Cas enzymology for specific recognition of desired DNA or RNA sequences. This platform, termed specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), allows multiplexed, portable, and ultra-sensitive detection of RNA or DNA from clinically relevant samples. Here, we provide step-by-step instructions for setting up SHERLOCK assays with recombinase-mediated polymerase pre-amplification of DNA or RNA and subsequent Cas13- or Cas12-mediated detection via fluorescence and colorimetric readouts that provide results in <1 h with a setup time of less than 15 min. We also include guidelines for designing efficient CRISPR RNA (crRNA) and isothermal amplification primers, as well as discuss important considerations for multiplex and quantitative SHERLOCK detection assays.





Similar content being viewed by others
Data and materials availability
Reagents from the Zhang lab are widely available to the academic community through Addgene, and additional information and resources can be found on the Zhang lab website (https://zlab.bio/) and GitHub (github.com/fengzhanglab).
Change history
31 January 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Piepenburg, O., Williams, C. H., Stemple, D. L. & Armes, N. A. DNA detection using recombination proteins. PLoS Biol. 4, e204 (2006).
Bhattacharyya, R. P., Thakku, S. G. & Hung, D. T. Harnessing CRISPR effectors for infectious disease diagnostics. ACS Infect. Dis. 4, 1278–1282 (2018).
Batista, A. C. & Pacheco, L. G. C. Detecting pathogens with Zinc-Finger, TALE and CRISPR-based pro grammable nucleic acid binding proteins. J. Microbiol. Methods 152, 98–104 (2018).
Li, Y., Li, S., Wang, J. & Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 37, 730–743 (2019).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Li, S. Y. et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493 (2018).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Smargon, A. A. et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018).
Yan, W. X. et al. Cas13d Is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e5 (2018).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017).
Spoelstra, W. K. et al. CRISPR-based DNA and RNA detection with liquid phase separation. Preprint at https://www.biorxiv.org/content/10.1101/471482v2 (2018).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Tomita, N., Mori, Y., Kanda, H. & Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 3, 877–882 (2008).
Vincent, M., Xu, Y. & Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 5, 795–800 (2004).
Neuzil, P., Zhang, C., Pipper, J., Oh, S. & Zhuo, L. Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 34, e77 (2006).
Farrar, J. S. & Wittwer, C. T. Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin. Chem. 61, 145–153 (2015).
Ameur, A., Kloosterman, W. P. & Hestand, M. S. Single-molecule sequencing: towards clinical applications. Trends Biotechnol. 37, 72–85 (2019).
Li, S. Y. et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 4, 20 (2018).
Li, L., Li, S. & Wang, J. CRISPR-Cas12b-assisted nucleic acid detection platform. Preprint at https://www.biorxiv.org/content/10.1101/362889v1 (2018).
Hajian, R. et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 3, 427–437 (2019).
Abudayyeh, O. O. et al. RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Kleinstiver, B. P. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448 (2018).
Niemz, A., Ferguson, T. M. & Boyle, D. S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 29, 240–250 (2011).
Compton, J. Nucleic acid sequence-based amplification. Nature 350, 91–92 (1991).
Guatelli, J. C. et al. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc. Natl Acad. Sci. 87, 1874–1878 (1990).
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 (2012).
East-Seletsky, A., O’Connell, M. R., Burstein, D., Knott, G. J. & Doudna, J. A. RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol. Cell 66, 373–383.e3 (2017).
Acknowledgements
We thank W. Blake and C. Brown for helpful feedback, and R. Macrae, R. Belliveau, E. Blackwell, and the entire Zhang lab for discussions and support. O.O.A. was supported by an NIH grant (F30 NRSA 1F30-CA210382). F.Z. was a New York Stem Cell Foundation–Robertson Investigator. F.Z. was supported by NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell and G. Harold and Leila Mathers foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and J. and P. Poitras, R. Metcalfe, and D. Cheng.
Author information
Authors and Affiliations
Contributions
M.J.K., J.G.K., J.S.G., O.O.A., and F.Z. designed and conducted all experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.J.K., J.S.G., O.O.A., and F.Z. are co-inventors on patent applications filed by the Broad Institute relating to work in this article. J.S.G., O.O.A., and F.Z. are co-founders of Sherlock Biosciences. F.Z. is a co-founder and advisor of Beam Therapeutics, Editas Medicine, Pairwise Plants, and Arbor Biotechnologies. O.O.A. and J.S.G. are advisers for Beam Therapeutics. J.S.G. is a campus advisor for Benchling, Inc.
Additional information
Peer review information Nature Protocols thanks Tetsushi Sakum, Jin Wang and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Gootenberg, J. S. et al. Science 356, 438−442 (2017): https://doi.org/10.1126/science.aam9321
Gootenberg, J. S. et al. Science 360, 439−444 (2018): https://doi.org/10.1126/science.aaq0179
Myhrvold, C. et al. Science 360, 444−448 (2018): https://doi.org/10.1126/science.aas8836
Supplementary information
Rights and permissions
About this article
Cite this article
Kellner, M.J., Koob, J.G., Gootenberg, J.S. et al. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc 14, 2986–3012 (2019). https://doi.org/10.1038/s41596-019-0210-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0210-2
- Springer Nature Limited
This article is cited by
-
Structures, mechanisms and applications of RNA-centric CRISPR–Cas13
Nature Chemical Biology (2024)
-
Ultrasensitive single-step CRISPR detection of monkeypox virus in minutes with a vest-pocket diagnostic device
Nature Communications (2024)
-
Do CRISPR-based disease diagnosis methods qualify as point-of-care diagnostics for plant diseases?
The Nucleus (2024)
-
CRISPR/Cas-mediated germplasm improvement and new strategies for crop protection
Crop Health (2024)
-
Carboxymethyl chitosan/alginate/polyethylene glycol composite hydrogel for nucleic acid lysis solution storage at the point of care
Cellulose (2024)