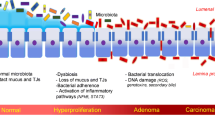
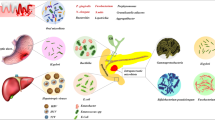
Abstract
Biliary tract cancers, including intra- and extra-hepatic cholangiocarcinoma as well as gallbladder cancer, are associated with poor prognosis and the majority of patients present with advanced-stage, non-resectable disease at diagnosis. Biliary tract cancer may develop through an accumulation of genetic and epigenetic alterations and can be influenced by microbial exposure. Furthermore, the liver and biliary tract are exposed to the gastrointestinal microbiome through the gut–liver axis. The availability of next-generation sequencing technology has led to an increase in studies investigating the relationship between microbiota and human disease. In particular, the interplay between the microbiome, the tumour micro-environment and response to systemic therapy is a prospering area of interest. Given the poor outcomes for patients with biliary tract cancer, this emerging field of research, through which new biomarkers may be identified, offers potential as a tool for early diagnosis, prognostication or even as a future therapeutic target. This review summarises the available evidence on the microbiome environment in patients with biliary tract cancer, including a discussion around confounding factors, implications for therapy and proposed future directions.

Similar content being viewed by others
References
Nakeeb A, Pitt HA, Sohn TA, Coleman JA, Abrams RA, Piantadosi S, et al. Cholangiocarcinoma: a spectrum of intrahepatic, perihilar, and distal tumors. Ann Surg. 1996;224:463–75.
Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261–80.
Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology. 2001;33:1353–7.
Taylor-Robinson SD, Toledano MB, Arora S, Keegan TJ, Hargreaves S, Beck A, et al. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968–1998. Gut. 2001;48:816–20.
Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open. 2021;4:e214708.
Forner A, Vidili G, Rengo M, Bujanda L, Ponz-Sarvisé M, Lamarca A. Clinical presentation, diagnosis and staging of cholangiocarcinoma. Liver Int. 2019;39:98–107.
Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–81.
Lamarca A, Palmer DH, Wasan HS, Ross PJ, Ma YT, Arora A, et al. Second-line FOLFOX chemotherapy versus active symptom control for advanced biliary tract cancer (ABC-06): a phase 3, open-label, randomised, controlled trial. Lancet Oncol. 2021;22:690–701.
Valle JW, Kelley RK, Nervi B, Oh DY, Zhu AX. Biliary tract cancer. Lancet. 2021;397:428–44.
Sithithaworn P, Yongvanit P, Duenngai K, Kiatsopit N, Pairojkul C. Roles of liver fluke infection as risk factor for cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014;21:301–8.
Clements O, Eliahoo J, Kim JU, Taylor-Robinson SD, Khan SA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J Hepatol. 2019;72:95–103.
Hsing AW, Bai Y, Andreotti G, Rashid A, Deng J, Chen J, et al. Family history of gallstones and the risk of biliary tract cancer and gallstones: A population-based study in Shanghai, China. Int J Cancer. 2007;121:832–8.
Lewis JT, Talwalkar JA, Rosen CB, Smyrk TC, Abraham SC. Prevalence and risk factors for gallbladder neoplasia in patients with primary sclerosing cholangitis: Evidence for a metaplasia-dysplasia-carcinoma sequence. Am J Surg Pathol. 2007;31:907–13.
Kamisawa T, Kaneko K, Itoi T, Ando H. Pancreaticobiliary maljunction and congenital biliary dilatation. Lancet Gastroenterol Hepatol. 2017;2:610–8.
Li ZM, Wu ZX, Han B, Mao YQ, Chen HL, Han SF, et al. The association between BMI and gallbladder cancer risk: a meta-analysis. Oncotarget. 2016;7:43669–79.
Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–10.
Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533.
Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84.
Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–9.
Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9.
Gao R, Wang Z, Li H, Cao Z, Gao Z, Chen H, et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J Gastroenterol Hepatol. 2020;35:2109–21.
Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: evidence of association and causality. Semin Immunol. 2017;32:25–34.
Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6.
Bäckhed F, Ding H, Wang T, Hooper LV, Gou YK, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23.
Ma HQ, Yu TT, Zhao XJ, Zhang Y, Zhang HJ. Fecal microbial dysbiosis in Chinese patients with inflammatory bowel disease. World J Gastroenterol. 2018;24:1464–77.
Wu SC, Cao ZS, Chang KM, Juang JL. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat Commun. 2017;8:24.
Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB, et al. Colonic bacterial composition in Parkinson’s disease. Mov Disord. 2015;30:1351–60.
Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411.
De Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–69.
Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22–39.
Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50.
Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72:558–77.
Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:eaan5931.
Scheufele F, Aichinger L, Jäger C, Demir IE, Schorn S, Sargut M, et al. Effect of preoperative biliary drainage on bacterial flora in bile of patients with periampullary cancer. Br J Surg. 2017;104:e182–e188.
Scheufele F, Schorn S, Demir IE, Sargut M, Tieftrunk E, Calavrezos L, et al. Preoperative biliary stenting versus operation first in jaundiced patients due to malignant lesions in the pancreatic head: a meta-analysis of current literature. Surg (U S). 2017;161:939–50.
Shrader HR, Miller AM, Tomanek-Chalkley A, McCarthy A, Coleman KL, Ear PH, et al. Effect of bacterial contamination in bile on pancreatic cancer cell survival. Surg (U S). 2021;169:617–22.
Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M, et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome. 2019;7:1–17.
Saab M, Mestivier D, Sohrabi M, Rodriguez C, Khonsari MR, Faraji A, et al. Characterization of biliary microbiota dysbiosis in extrahepatic cholangiocarcinoma. PLoS ONE. 2021;16:e0247798.
Xiao M, Gao Y, Wang Y. Helicobacter species infection may be associated with cholangiocarcinoma: a meta‐analysis. Int J Clin Pr. 2014;68:262–70.
Zhou D, Wang JD, Weng MZ, Zhang Y, Wang XF, Gong W, et al. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: A meta-analysis. Eur J Gastroenterol Hepatol. 2013;25:447–54.
Segura-López FK, Avilés-Jiménez F, Güitrón-Cantú A, Valdéz-Salazar HA, León-Carballo S, Guerrero-Pérez L, et al. Infection with Helicobacter bilis but not Helicobacter hepaticus was Associated with Extrahepatic Cholangiocarcinoma. Helicobacter. 2015;20:223–30.
Avilés-Jiménez F, Guitron A, Segura-López F, Méndez-Tenorio A, Iwai S, Hernández-Guerrero A, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect. 2016;22:178.
Murphy G, Michel A, Taylor PR, Albanes D, Weinstein SJ, Virtamo J, et al. Association of seropositivity to Helicobacter species and biliary tract cancer in the ATBC study. Hepatology. 2014;60:1963–71.
Nagaraja V, Eslick GD. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall‐bladder cancer. Aliment Pharm Ther. 2014;39:745–50.
Koshiol J, Wozniak A, Cook P, Adaniel C, Acevedo J, Azócar L, et al. Salmonella enterica serovar Typhi and gallbladder cancer: a case-control study and meta-analysis. Cancer Med. 2016;5:3310–25.
Serra N, Di Carlo P, Gulotta G, d’Arpa F, Giammanco A, Colomba C, et al. Bactibilia in women affected with diseases of the biliary tract and pancreas. A STROBE guidelines-adherent cross-sectional study in Southern Italy. J Med Microbiol. 2018;67:1090–5.
Di Carlo P, Serra N, D’arpa F, Agrusa A, Gulotta G, Fasciana T, et al. The microbiota of the bilio-pancreatic system: a cohort, STROBE-compliant study. Infect Drug Resist. 2019;12:1513–27.
Tsuchiya Y, Loza E, Villa-Gomez G, Trujillo CC, Baez S, Asai T, et al. Metagenomics of microbial communities in gallbladder bile from patients with gallbladder cancer or cholelithiasis. Asian Pacific. J Cancer Prev. 2018;19:961–7.
Poudel SK, Padmanabhan R, Chahal P, Sanaka M, Stevens T, Guinta K, et al. Microbiome signature of bile from pancreatic and biliary tract cancer patients: A pilot study. J Clin Oncol. 2019;37:e15744–e15744.
Jia X, Lu S, Zeng Z, Liu Q, Dong Z, Chen Y, et al. Characterization of gut microbiota, bile acid metabolism, and cytokines in intrahepatic cholangiocarcinoma. Hepatology. 2020;71:893–906.
Chng KR, Chan SH, Ng AHQ, Li C, Jusakul A, Bertrand D, et al. Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine. 2016;8:195–202.
Chen B, Fu SW, Lu L, Zhao H. A preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma. Biomed Res Int. 2019;1092563.
Dangtakot R, Intuyod K, Ahooja A, Wongwiwatchai J, Hanpanich P, Lulitanond A, et al. Profiling of bile microbiome identifies district microbial population between choledocholithiasis and cholangiocarcinoma patients. Asian Pac J Cancer Prev. 2021;22:233–40.
Zhang L, Wu YN, Chen T, Ren CH, Li X, Liu GX. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat Dis Int. 2019;18:149–57.
Lenz P, Steidl L, Cordes F, Kahl BC, Karch H, Dobrindt U, et al. Sa1328 analysis of the human biliary microbiome and its alterations in biliary tract diseases. Gastroenterology. 2015;148:S–293.
Katsuyuki M, Chandrasekhara V, Wongjarupong N, Chen J, Johnson S, Chia N, et al.The Role of the Biliary and Gut Microbiome in the Progression of Cholangiocarcinoma and Primary Sclerosing Cholangitis. Am J Gastroenterol. 2019;114:S25–6.
Lee H, Lee HK, Min SK, Lee WH. 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer. World J Surg Oncol. 2020;18:19.
Song X, Wang X, Hu Y, Li H, Ren T, Li Y, et al. A metagenomic study of biliary microbiome change along the cholecystitis‐carcinoma sequence. Clin Transl Med. 2020;10:e97.
Voigt AY, Costea PI, Kultima JR, Li SS, Zeller G, Sunagawa S, et al. Temporal and technical variability of human gut metagenomes. Genome Biol. 2015;16:73.
Galloway-Peña JR, Smith DP, Sahasrabhojane P, Wadsworth WD, Fellman BM, Ajami NJ, et al. Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients. Genome Med. 2017;9:21.
He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24:1532–5.
Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531–531.
Langheinrich M, Wirtz S, Kneis B, Gittler MM, Tyc O, Schierwagen R, et al. Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development. J Clin Med. 2020;9:2785.
Wang Z, Zolnik CP, Qiu Y, Usyk M, Wang T, Strickler HD, et al. Comparison of fecal collection methods for microbiome and metabolomics studies. Front Cell Infect Microbiol. 2018;8:301.
Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, et al. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems. 2016;1:21–37.
Wu WK, Chen CC, Panyod S, Chen RA, Wu MS, Sheen LY, et al. Optimization of fecal sample processing for microbiome study—the journey from bathroom to bench. J Formos Med Assoc. 2019;118:545–55.
Gupta S, Mortensen MS, Schjørring S, Trivedi U, Vestergaard G, Stokholm J, et al. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun Biol. 2019;2:1–7.
Woo PCY, Lau SKP, Teng JLL, Tse H, Yuen KY. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14:908–34.
Claassen S, du Toit E, Kaba M, Moodley C, Zar HJ, Nicol MP. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J Microbiol Methods. 2013;94:103–10.
Pollock J, Glendinning L, Wisedchanwet T, Watson M. The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies. Appl Environ Microbiol. 2018;84:e02627–17.
Guo F, Zhang T. Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl Microbiol Biotechnol. 2013;97:4607–16.
Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinforma. 2016;17:135.
Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, et al. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6:771.
Bergsten E, Mestivier D, Sobhani I. The limits and avoidance of biases in metagenomic analyses of human fecal microbiota. Microorganisms. 2020;8:1954.
Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7.
Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103.
Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–8.
Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806.
Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–60.
McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS ONE. 2013;8:e53653.
Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227–33.
Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–68.
Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–8.
Pereira P, Aho V, Arola J, Boyd S, Jokelainen K, Paulin L, et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS ONE. 2017;12:e0182924.
Lurienne L, Cervesi J, Duhalde L, de Gunzburg J, Andremont A, Zalcman G, et al. NSCLC immunotherapy efficacy and antibiotic use: a systematic review and meta-analysis. J Thorac Oncol. 2020;15:1147–59.
Wilson BE, Routy B, Nagrial A, Chin VT. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: a systematic review and meta-analysis of observational studies. Cancer Immunol Immunother. 2020;69:343–54.
Petrelli F, Iaculli A, Signorelli D, Ghidini A, Dottorini L, Perego G, et al. Survival of patients treated with antibiotics and immunotherapy for cancer: a systematic review and meta-analysis. J Clin Med. 2020;9:1458.
Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–9.
Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: Insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15:1–20.
Brennan CA, Garrett WS. Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–66.
Zhao H, Chu M, Huang Z, Yang X, Ran S, Hu B, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7:11773.
Harrandah AM, Chukkapalli SS, Bhattacharyya I, Progulske-Fox A, Chan EKL. Fusobacteria modulate oral carcinogenesis and promote cancer progression. J Oral Microbiol. 2021;13:1849493.
Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22:5574–81.
Liu Y, Baba Y, Ishimoto T, Tsutsuki H, Zhang T, Nomoto D, et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer. 2021;124:963–74.
Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS ONE. 2016;11:e0153274.
Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased Abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8:158.
Boehm ET, Thon C, Kupcinskas J, Steponaitiene R, Skieceviciene J, Canbay A, et al. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci Rep. 2020;10:16240.
Yurdakul D, Yazgan-Karataş A, Şahin F. Enterobacter strains might promote colon cancer. Curr Microbiol. 2015;71:403–11.
Acknowledgements
Not applicable
Funding
Dr. Roseanna C Wheatley is studying for an MD, funded by the Timpson Fellowship. The salary of Dr. Angela Lamarca is in part funded by The Christie Charity and the European Union’s Horizon 2020 Research and Innovation Programme [grant number 825510, ESCALON].
Author information
Authors and Affiliations
Contributions
RW drafted and revised the paper. EK reviewed and approved the final version of the paper. TJ performed the literature search and reviewed and approved the final version of the paper. AL reviewed and approved the final version of the paper. RAH reviewed and approved the final version of the paper. JWV reviewed and approved the final version of the paper. MMN concept initialisation and review and approval of draft and final version of the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
RCW: no competing interests to declare. EK: no competing interests to declare. TJ: no competing interests to declare. AL: received travel and educational support from Ipsen, Pfizer, Bayer, AAA, SirtEx, Novartis, Mylan and Delcath; speaker honoraria from Merck, Pfizer, Ipsen, Incyte and AAA; advisory honoraria from EISAI, Nutricia Ipsen, QED and Roche; she is a member of the Knowledge Network and NETConnect Initiatives funded by Ipsen; all outside scope of this work. RAH has served on the advisory board for Roche, BMS, Eisai, Celgene, Beigene, Ipsen and BTG. He has received speaker fees from Eisai, Ipsen, Mylan and PrimeOncology, and has received travel and educational support from Bayer, BMS and Roche; all outside of the scope of this work. JWV received honoraria from Agios, AstraZeneca, Baxter, Genoscience Pharma, Hutchison Medipharma, Imaging Equipment Ltd (AAA), Incyte, Ipsen, Mundipharma EDO, Mylan, QED, Servier, Sirtex and Zymeworks; and grants, honoraria and non-financial support from NuCana, all outside of the scope of this work. MMN received research grant support from Servier, Ipsen and NuCana. She has received travel and accommodation support from Bayer and Ipsen and speaker honoraria from Pfizer, Ipsen, NuCana and Mylan. She has served on advisory boards for Celgene, Ipsen, Sirtex, Baxalta and Incyte; all outside of the scope of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wheatley, R.C., Kilgour, E., Jacobs, T. et al. Potential influence of the microbiome environment in patients with biliary tract cancer and implications for therapy. Br J Cancer 126, 693–705 (2022). https://doi.org/10.1038/s41416-021-01583-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01583-8
- Springer Nature Limited