Abstract
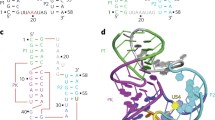
The crystal structure of the RNA fragment, 5′-r(UUCGCG)-3′, has been determined at 1.4 Å resolution by a combination of single isomorphous replacement and molecular search methods. The 3′-terminal CGCG portion of the hexamer engages in Watson–Crick hydrogen bonding while the S′-terminal UU-overhang forms novel Hoogsteen-like UU self-base pairs with the overhang of an adjacent duplex. The U·U pairs display a single conventional hydrogen bond between O4 (U1) and N3 (U8) and a CH–O hydrogen bond between C5-H (U1) and O4(U8), through the Hoogsteen face of the pyrimidine base U1. This unusual arrangement of one of the bases results in a trans U·U pair on antiparallel strands in contrast to the usual cis base pairs. The structure emphasizes the pronounced polymorphism of U·U pairs and has implications for folding of RNA molecules.
Similar content being viewed by others
References
Pley, H.W., Flaherty, K.M. & McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74 (1994).
Scott, W.G., Finch, J.T. & Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81, 991–1002 (1995).
Michel, F. & Westhof, E. Modeling of the three-dimensional architecture of group I catalytic introns on comparative sequence analysis. J. molec. Biol. 216, 585–610 (1990).
Holbrook, S.R.C., Tinoco Jr., I.S Kim, S. H. Crystal structure of an RNA double helix incorporating a track of non-Watson–Crick base pairs. Nature 353, 579–581 (1991).
Baeyens, K.J., De Bondt, H.L. & Holbrook, S.R. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nature struct. Biol. 2, 56–62 (1995).
Bhattacharyya, D. & Bansal, M. Local variability and base sequence effects in DNA crystal structures. J. Biomol. struct. Dyn. 8, 539–572 (1990).
Calladine, C.R. Mechanism of sequence-dependent stacking of bases in B-DNA. J. molec. Biol. 161, 343–352 (1982).
Westhof, E. & Sundaralingam, M. X-ray structure of a cytidyl-3′,5′- adenosine-proflavine complex: A self-paired parallel-chain double helical dimer with an intercalated acridine dye. Proc. natn. Acad. Sci. U.S.A. 77, 1852–1856 (1980).
Kang, C.H. et al. Crystal structure of intercalated four-stranded d(C3T) at 1.4A resolution. Proc. natn. Acad. Sci. U.S.A. 91, 11636–11640 (1994).
Westhof, E. Westhof's rule. Nature 358, 459–460 (1992).
Lavery, R., Zakrezewska, K., Sun, J.S. & Harvey, S.C. A comprehensive classification of nucleic acid structural families based on strand direction and base pairing. Nucleic Acids Res. 20, 5011–5016 (1992).
Leonard, G.A., McAuley-Hecht, K.E., Brown, T. & Hunter, W.N. Do CH-O hydrogen bonds contribute to the stability of nucleic acid base pairs? Acta Crystallogr. D51, 136–139 (1995).
Derewenda, Z.S., Lee, L. & Derewenda, U. The occurrence of C-H–O hydrogen bonds in proteins. J. molec. Biol. 252, 248–262 (1995).
Michel, F., Ellington, A.D., Couture, S. & Szostak, J. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature 347, 578–580 (1990).
Cruse, W. et al. The structure of a mispaired RNA double helix at 1.6Å resolution and implications for the prediction of RNA secondary structure. Proc. natn. Acad. Sci. U.S.A. 91, 4160–4164 (1994).
Wang, A.H.J. et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686 (1979).
Bugg, C.E., Thomas, J.M., Sundaralingam, M. & Rao, S.T. Stereochemistry of nucleic acids and their constituents. X. Solid-state base-stacking patterns in nucleic acid constituents and polynucleotides. Biopolymers 10, 175–219 (1971).
Betzel, C., Lorenz, S., Fiirste, J.P., Bald, R., Zhang, M., Schneider, T.R., Wilson, K.S. & Erdmann, V.A. Crystal structure of domain A of Thermus Flavus 55 rRNA and the contribution of water molecules to its structure. FEBSLett. 351, 1509–164 (1994).
Hall, K., Cruz, P., Tinoco Jr, I., Jovin, T. & van de Sande, J. H. ‘Z-RNA’ - a left-handed RNA double helix. Nature 311, 584–586 (1984).
Westhof, E. & Michel, F. Prediction and experimental investigation of RNA secondary and tertiary foldings, in RNA-protein interactions (eds K. Nagai, & I. W. Mattaj) 25–51 (Oxford University Press Inc., New York, NY, 1994).
van Meervelt, L. et al. High-resolution structure of a DNA helix forming (C–G)*G base triplets. Nature 374, 742–744 (1995).
Howard, A.J., Nielsen, C. & Xuang, N.H. Software for a diffractometer with multiwire area detector. Meths. Enzymol. 144, 211–237 (1985).
Wang, B.C. Resolution of phase ambiguity in macromolecular crystallography. Meths. Enzymol. 115, 90–112 (1985).
Brünger, A.T. X-PLOR - A system for x-ray crystallography and NMR (Yale University Press, New Haven, CT, 1992).
Sack, J. & Quiocho, F.A. CHAIN - Crystallographic modeling program (Baylor College of Medicine, Houston, TX, 1992).
Brunger, A.T. Crystallographic refinement by simulated annealing, in Crystallographic computing 4: Techniques and new technologies (eds N. W.Isaacs, & M.R. Taylor) 126–140 (Clarendon Press, Oxford, 1988).
Bernstein, F.C. et al. The protein data bank: a computer-based archival file for macromolecule structures. J. molec. Biol. 112, 535–542 (1977).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Wahl, M., Rao, S. & Sundaralingam, M. The structure of r(UUCGCG) has a 5′-UU-overhang exhibiting Hoogsteen-like trans U•U base pairs. Nat Struct Mol Biol 3, 24–31 (1996). https://doi.org/10.1038/nsb0196-24
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb0196-24
- Springer Nature America, Inc.
This article is cited by
-
A New Perspective on the Maillard Reaction and the Origin of Life
Journal of Molecular Evolution (2021)
-
Exploring the C–H…O Interactions in Glycoproteins
Applied Biochemistry and Biotechnology (2009)
-
Theoretical studies on the properties of uracil and its dimer upon thioketo substitution
Theoretical Chemistry Accounts (2008)
-
Interguanine hydrogen-bonding patterns in adducts with water and Zn–purine complexes (purine is 9-methyladenine and 9-methylguanine). Unexpected preference of Zn(II) for adenine-N7 over guanine-N7
JBIC Journal of Biological Inorganic Chemistry (2007)
-
Conformational specificity of non-canonical base pairs and higher order structures in nucleic acids: crystal structure database analysis
Journal of Computer-Aided Molecular Design (2006)