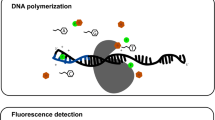
Abstract
We describe a protocol for construction and quantification of libraries for emulsion PCR (emPCR)-based sequencing platforms such as Roche 454 or Ion Torrent PGM. The protocol involves library construction using customized Y adapters, quantification using TaqMan-MGB (minor groove binder) probe–based quantitative PCR (qPCR) and calculation of an optimal template-to-bead ratio based on Poisson statistics, thereby avoiding the need for a laborious titration assay. Unlike other qPCR methods, the TaqMan-MGB probe specifically quantifies effective libraries in molar concentration and does not require specialized equipment. A single quality control step prior to emulsion PCR ensures that libraries contain no adapter dimers and have an optimal length distribution. The presented protocol takes ∼7 h to prepare eight barcoded libraries from genomic DNA into libraries that are ready to use for full-scale emPCR. It will be useful, for example, to allow analyses of precious clinical samples and amplification-free metatranscriptomics.




Similar content being viewed by others
References
Roche Diagnostics. Rapid Library Preparation Method Manual: GS FLX Titanium Series (Roche Applied Science, October 2009 (Rev. January 2010)).
Frias-Lopez, J. et al. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA 105, 3805–3810 (2008).
Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12, R18 (2011).
Davey, J.L. & Blaxter, M.W. RADSeq: next-generation population genetics. Brief Funct. Genomics 9, 416–423 (2010).
Zheng, Z. et al. Titration-free massively parallel pyrosequencing using trace amounts of starting material. Nucleic Acids Res. 38, e137 (2010).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Baird, N.A. et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 (2008).
Carson, S. et al. DNA sequencing by capillary electrophoresis: use of a two-laser-two-window intensified diode array detection system. Anal. Chem. 65, 3219–3226 (1993).
White, R.A. III, Blainey, P.C., Fan, H.C. & Quake, S.R. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics 10, 116 (2009).
Huang, J., Zheng, Z., Andersen, A.F., Engstran, L. & Ye, W. Rapid screening of complex DNA samples by single-molecule amplification and sequencing. PLoS ONE 6, e19723 (2011).
Roche Diagnostics. emPCR Method Manual: Lib-L LV GS FLX Titanium Series (Roche Applied Science, October 2009 (Rev. January 2010)).
Roche Diagnostics. Sequencing Method Manual: GS FLX Titanium Series (Roche Applied Science, October 2009 (Rev. November 2010)).
Acknowledgements
This study was supported by the Sixth Research Framework Programme of the European Union, a project on infections and cancers (INCA, LSHC-CT-2005-018704); the Karolinska Institutet Faculty funds for partial financing of new doctoral students (KID scholarship) to Z.Z.; and the Swedish Research Council Formas grant (FORMAS) to A.F.A.
Author information
Authors and Affiliations
Contributions
Z.Z. designed the Y adapters and qPCR assay, proposed Poisson statistics, conducted the experiments and drafted the manuscript; A.A., Ö.M., S.G. and H.N. assisted with the experiments and provided critical reviews; W.Y., L.E. and A.A. obtained funding and provided critical reviews; A.F.A. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1
454 Titanium library pooling calculation.xls: An example of calculation for 454 sample library pooling. (XLS 41 kb)
Supplementary Data 2
TrimYadapter.txt: A template file for user-customized Y adapter trimming. (XML 5 kb)
Supplementary Data 3
Yscheme.txt: User-defined MID sets for the 8 Y adapters. (TXT 0 kb)
Rights and permissions
About this article
Cite this article
Zheng, Z., Advani, A., Melefors, Ö. et al. Titration-free 454 sequencing using Y adapters. Nat Protoc 6, 1367–1376 (2011). https://doi.org/10.1038/nprot.2011.369
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.369
- Springer Nature Limited
This article is cited by
-
Prediction of smoking by multiplex bisulfite PCR with long amplicons considering allele-specific effects on DNA methylation
Clinical Epigenetics (2018)
-
Regulation of PfEMP1–VAR2CSA translation by a Plasmodium translation-enhancing factor
Nature Microbiology (2017)
-
Unlocking the mystery of the hard-to-sequence phage genome: PaP1 methylome and bacterial immunity
BMC Genomics (2014)
-
Consistency of metagenomic assignment programs in simulated and real data
BMC Bioinformatics (2014)
-
Anchored multiplex PCR for targeted next-generation sequencing
Nature Medicine (2014)