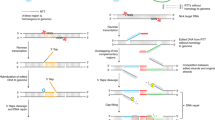
Abstract
Targeted DNA double-strand breaks introduced by rare-cleaving designer endonucleases can be harnessed for gene disruption applications by engaging mutagenic nonhomologous end-joining DNA repair pathways. However, endonuclease-mediated DNA breaks are often subject to precise repair, which limits the efficiency of targeted genome editing. To address this issue, we coupled designer endonucleases to DNA end–processing enzymes to drive mutagenic break resolution, achieving up to 25-fold enhancements in gene disruption rates.



Similar content being viewed by others
References
Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. & Gregory, P.D. Nat. Rev. Genet. 11, 636–646 (2010).
Lieber, M.R. Annu. Rev. Biochem. 79, 181–211 (2010).
McVey, M. & Lee, S.E. Trends Genet. 24, 529–538 (2008).
Mladenov, E. & Iliakis, G. Mutat. Res. 711, 61–72 (2011).
Shrivastav, M., De Haro, L.P. & Nickoloff, J.A. Cell Res. 18, 134–147 (2008).
Bennardo, N., Gunn, A., Cheng, A., Hasty, P. & Stark, J.M. PLoS Genet. 5, e1000683 (2009).
Szymczak, A.L. & Vignali, D.A.A. Expert Opin. Biol. Ther. 5, 627–638 (2005).
Certo, M.T. et al. Nat. Methods 8, 671–676 (2011).
Maeder, M.L., Thibodeau-Beganny, S., Sander, J.D., Voytas, D.F. & Joung, J.K. Nat. Protoc. 4, 1471–1501 (2009).
Miller, J.C. et al. Nat. Biotechnol. 29, 143–148 (2011).
Cannon, P. & June, C. Curr. Opin. HIV AIDS 6, 74–79 (2011).
Smith, J. et al. Nucleic Acids Res. 34, e149 (2006).
Pattanayak, V., Ramirez, C.L., Joung, J.K. & Liu, D.R. Nat. Methods 8, 765–770 (2011).
Sather, B.D. et al. Mol. Ther. 19, 515–525 (2011).
Cermak, T. et al. Nucleic Acids Res. 39, e82 (2011).
Acknowledgements
M.T.C. was supported in part by the US Public Health Service and National Research Service Award (T32 GM07270) from the US National Institute of General Medical Sciences. Additional funding was from the US National Institutes of Health (RL1CA133832, UL1DE019582, R01-HL075453, PL1-HL092557, RL1-HL092553, RL1-HL92554 and U19-AI96111) and Seattle Children's Center for Immunity and Immunotherapies. We would like to thank J. Stark (Beckman Research Institute and City of Hope) and D. Stetson (University of Washington) for initial Trex2 expression plasmids and D. Voytas (University of Minnesota) for TALEN cloning reagents. Zinc-finger nuclease expression plasmids were provided by C. Ramirez and K. Joung (Harvard University and Massachusetts General Hospital). J. Jarjour (Seattle Children's Research Institute, presently Pregenen Inc.) designed the Sce-SCID mouse, and Sce-SCID MEFs were provided by A. Astrakhan and G. Metzler (University of Washington). We thank all members of the Northwest Genome Editing Consortium for their many insightful discussions.
Author information
Authors and Affiliations
Contributions
M.T.C. designed and conceived experiments. M.T.C., K.S.G., R.K., B.S., G.C., M.B. and T.M. performed experiments. A.R.L., S.K.B., K.J., M.B., B.Y.R., H.-P.K., A.G. and F.P. provided reagents. M.T.C. wrote the paper and K.S.G., D.J.R. and A.M.S. edited the paper. D.J.R. and A.M.S provided technical support and conceptual advice.
Corresponding authors
Ethics declarations
Competing interests
A.M.S., F.P. and A.G. are employees of Cellectis and receive salary and equity compensation. A.M.S. is cofounder and board member of Pregenen Inc., a genome engineering company. A provisional patent (no. 61/447,672) for coupling endonucleases with exonucleases for gene disruption has been filed by Seattle Children's Research Institute.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1 and 2 and Supplementary Note 1 (PDF 1352 kb)
Rights and permissions
About this article
Cite this article
Certo, M., Gwiazda, K., Kuhar, R. et al. Coupling endonucleases with DNA end–processing enzymes to drive gene disruption. Nat Methods 9, 973–975 (2012). https://doi.org/10.1038/nmeth.2177
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2177
- Springer Nature America, Inc.
This article is cited by
-
Enhanced genome editing efficiency of CRISPR PLUS: Cas9 chimeric fusion proteins
Scientific Reports (2021)
-
Yeast genetic interaction screens in the age of CRISPR/Cas
Current Genetics (2019)
-
Biased genome editing using the local accumulation of DSB repair molecules system
Nature Communications (2018)
-
Molecular Evidence of Genome Editing in a Mouse Model of Immunodeficiency
Scientific Reports (2018)
-
Double deficiency of Trex2 and DNase1L2 nucleases leads to accumulation of DNA in lingual cornifying keratinocytes without activating inflammatory responses
Scientific Reports (2017)