Abstract
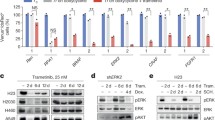
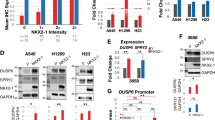
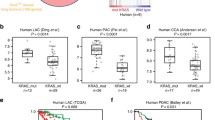
Patients with advanced Kirsten rat sarcoma viral oncogene homolog (KRAS)-mutant lung adenocarcinoma are currently treated with standard chemotherapy because of a lack of efficacious targeted therapies. We reasoned that the identification of mediators of Kras signaling in early mouse lung hyperplasias might bypass the difficulties that are imposed by intratumor heterogeneity in advanced tumors, and that it might unveil relevant therapeutic targets. Transcriptional profiling of KrasG12V-driven mouse hyperplasias revealed intertumor diversity with a subset that exhibited an aggressive transcriptional profile analogous to that of advanced human adenocarcinomas. The top-scoring gene in this profile encodes the tyrosine kinase receptor DDR1. The genetic and pharmacological inhibition of DDR1 blocked tumor initiation and tumor progression, respectively. The concomitant inhibition of both DDR1 and Notch signaling induced the regression of KRAS;TP53-mutant patient-derived lung xenografts (PDX) with a therapeutic efficacy that was at least comparable to that of standard chemotherapy. Our data indicate that the combined inhibition of DDR1 and Notch signaling could be an effective targeted therapy for patients with KRAS-mutant lung adenocarcinoma.






Similar content being viewed by others
References
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Pikor, L.A., Ramnarine, V.R., Lam, S. & Lam, W.L. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 82, 179–189 (2013).
Meng, D. et al. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: a systematic review with meta-analysis. Lung Cancer 81, 1–10 (2013).
Peters, S., Zimmermann, S. & Adjei, A.A. Oral epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of non-small cell lung cancer: comparative pharmacokinetics and drug-drug interactions. Cancer Treat. Rev. 40, 917–926 (2014).
Gridelli, C. et al. ALK inhibitors in the treatment of advanced NSCLC. Cancer Treat. Rev. 40, 300–306 (2014).
Jänne, P.A. et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 14, 38–47 (2013).
Oxnard, G.R., Binder, A. & Jänne, P.A. New targetable oncogenes in non-small-cell lung cancer. J. Clin. Oncol. 31, 1097–1104 (2013).
Pirker, R. Novel drugs against non-small-cell lung cancer. Curr. Opin. Oncol. 26, 145–151 (2014).
Reck, M. et al. LUME-Lung 1 Study Group. Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non-small-cell lung cancer (LUME-Lung 1): a phase 3, double-blind, randomised controlled trial. Lancet Oncol. 15, 143–155 (2014).
Gerlinger, M. et al. Cancer: evolution within a lifetime. Annu. Rev. Genet. 48, 215–236 (2014).
Burrell, R.A. & Swanton, C. The evolution of the unstable cancer genome. Curr. Opin. Genet. Dev. 24, 61–67 (2014).
Sweet-Cordero, A. et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet. 37, 48–55 (2005).
Valiathan, R.R., Marco, M., Leitinger, B., Kleer, C.G. & Fridman, R. Discoidin domain receptor tyrosine kinases: new players in cancer progression. Cancer Metastasis Rev. 31, 295–321 (2012).
Valencia, K. et al. Inhibition of collagen receptor discoidin domain receptor-1 (DDR1) reduces cell survival, homing, and colonization in lung cancer bone metastasis. Clin. Cancer Res. 18, 969–980 (2012).
Yang, S.H. et al. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung carcinomas. Oncol. Rep. 24, 311–319 (2010).
Miao, L. et al. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung cancer and promotes cell invasion via epithelial-to-mesenchymal transition. Med. Oncol. 30, 626 (2013).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Guerra, C. et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4, 111–120 (2003).
Monti, S., Tamayo, P., Mesirov, J. & Golub, T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 52, 91–118 (2003).
Okayama, H. et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–111 (2012).
Sánchez, M.P. et al. Multiple tyrosine protein kinases in rat hippocampal neurons: isolation of Ptk-3, a receptor expressed in proliferative zones of the developing brain. Proc. Natl. Acad. Sci. USA 91, 1819–1823 (1994).
Leitinger, B. Discoidin domain receptor functions in physiological and pathological conditions. Int. Rev. Cell Mol. Biol. 310, 39–87 (2014).
Vogel, W.F., Aszódi, A., Alves, F. & Pawson, T. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol. Cell. Biol. 21, 2906–2917 (2001).
Guerrot, D. et al. Discoidin domain receptor 1 is a major mediator of inflammation and fibrosis in obstructive nephropathy. Am. J. Pathol. 179, 83–91 (2011).
Gao, M. et al. Discovery and optimization of 3-(2-(Pyrazolo[1,5-a]pyrimidin-6-yl)ethynyl)benzamides as novel selective and orally bioavailable discoidin domain receptor 1 (DDR1) inhibitors. J. Med. Chem. 56, 3281–3295 (2013).
Kim, H.-G., Hwang, S.-Y., Aaronson, S.A., Mandinova, A. & Lee, S.W. DDR1 receptor tyrosine kinase promotes prosurvival pathway through Notch1 activation. J. Biol. Chem. 286, 17672–17681 (2011).
Weijzen, S. et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8, 979–986 (2002).
Maraver, A. et al. Therapeutic effect of γ-secretase inhibition in KrasG12V-driven non-small cell lung carcinoma by derepression of DUSP1 and inhibition of ERK. Cancer Cell 22, 222–234 (2012).
Licciulli, S. et al. Notch1 is required for Kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer Res. 73, 5974–5984 (2013).
Ito, T. et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127, 3913–3921 (2000).
Chen, Z. et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 483, 613–617 (2012).
Hegde, G.V. et al. Blocking NRG1 and other ligand-mediated Her4 signaling enhances the magnitude and duration of the chemotherapeutic response of non-small cell lung cancer. Sci. Transl. Med. 5, 171ra18 (2013).
Westhoff, B. et al. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA 106, 22293–22298 (2009).
Hassan, K.A. et al. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clin. Cancer Res. 19, 1972–1980 (2013).
Zheng, Y. et al. A rare population of CD24+ITGB4+Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell 24, 59–74 (2013).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Li, J. et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 6, 291–299 (2010).
Crystal, A.S. et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346, 1480–1486 (2014).
Bailis, W., Yashiro-Ohtani, Y. & Pear, W.S. Identifying direct Notch transcriptional targets using the GSI-washout assay. Methods Mol. Biol. 1187, 247–254 (2014).
Meacham, C.E. & Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Mainardi, S. et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc. Natl. Acad. Sci. USA 111, 255–260 (2014).
Desai, T.J., Brownfield, D.G. & Krasnow, M.A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014).
Gould, S.E., Junttila, M.R. & de Sauvage, F.J. Translational value of mouse models in oncology drug development. Nat. Med. 21, 431–439 (2015).
Day, C.-P., Merlino, G. & Van Dyke, T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 163, 39–53 (2015).
Johnson, F.M. et al. Phase II study of dasatinib in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 28, 4609–4615 (2010).
Blasco, R.B. et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011).
Karreth, F.A., Frese, K.K., DeNicola, G.M., Baccarini, M. & Tuveson, D.A. C-Raf is required for the initiation of lung cancer by K-Ras(G12D). Cancer Discov. 1, 128–136 (2011).
Chetoui, N., El Azreq, M.-A., Boisvert, M., Bergeron, M.-È. & Aoudjit, F. Discoidin domain receptor 1 expression in activated T cells is regulated by the ERK MAP kinase signaling pathway. J. Cell. Biochem. 112, 3666–3674 (2011).
Yen, W.C. et al. Anti-DLL4 has broad spectrum activity in pancreatic cancer dependent on targeting DLL4-Notch signaling in both tumor and vasculature cells. Clin. Cancer Res. 18, 5374–5386 (2012).
Andersson, E.R. & Lendahl, U. Therapeutic modulation of Notch signalling—are we there yet? Nat. Rev. Drug Discov. 13, 357–378 (2014).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Hoffman, R.M. Clinically accurate orthotopic mouse models of cancer. Methods Mol. Med. 58, 251–275 (2001).
Ambrogio, C. et al. Modeling lung cancer evolution and preclinical response by orthotopic mouse allografts. Cancer Res. 74, 5978–5988 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics 26, 589–595 (2010).
Koboldt, D.C. et al. VarScan2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Smyth, G.K., Michaud, J. & Scott, H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075 (2005).
Rothe, C. et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J. Mol. Biol. 376, 1182–1200 (2008).
Puyol, M. et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 18, 63–73 (2010).
Acknowledgements
We thank T. Hoey (OncoMed Pharmaceuticals) for providing us with demcizumab and for critical analysis of the data, as well as A. Vivancos (VHIO) for MiSeq analysis. We are thankful to S. Lee (Massachusetts General Hospital and Harvard University) for advice and reagents. We are grateful to O. Kocher, R. Pal and M. Joseph (Beth Israel Deaconess Medical Center) for help with LCM. This work was supported by grants from the European Research Council (ERC-AG/250297-RAS AEAD), the EU-Framework Programme (LSHG-CT-2007-037665/CHEMORES; HEALTH-F2-2010-259770/LUNGTARGET; and HEALTH-2010-260791/EUROCANPLATFORM), the Spanish Ministry of Economy and Competitiveness (SAF2011-30173 and SAF2014-59864-R), the Autonomous Community of Madrid (S2011/BDM-2470/ONCOCYCLE) and an Endowed Chair from the AXA Research Fund (all to M.B.), Fondo de Investigaciones Sanitarias FIS (PI13-01339 and Oncoprofile) and Fundación Mutua Madrileña AP150932014 (to A.Villanueva), a Juan Rodés fellowship from the Carlos III Health Institute (JR13/0002) and Fondo de Investigaciones Sanitarias (FIS PI14-01109) (to E.N.), the National Science Fund for Distinguished Young Scholars of China (81425021; to K.D.) and a postdoctoral fellowship from the Spanish Association Against Cancer, AECC (to C.A.). We thank R. Chiarle, T. Cash, S. Marro, M.L. De Bonis and A. Maraver for their critical reading of the manuscript and L. Paz-Ares for constructive discussion.
Author information
Authors and Affiliations
Contributions
C.A., D.S., A.Villanueva, M.S., M.H. and M.B. designed experiments, analyzed data and wrote the manuscript. C.A., M.F., A.Villanueva, P.J.F.-M., N.C. and R.B.B. performed experiments and analyzed the data. G.G.-L. performed all bioinformatic studies and did analysis. K.D., X.R. and Z.W. developed and provided the DDR1 inhibitor 7rh. M.S.-C. provided clinical samples and advice. A.Vidal performed human histopathological analysis. E.N. generated and evaluated clinical data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Tables 1 and 3 (PDF 20203 kb)
Supplementary Table 2
Significantly enriched gene pathways found in H2 versus H1 signatures. (XLSX 17 kb)
Rights and permissions
About this article
Cite this article
Ambrogio, C., Gómez-López, G., Falcone, M. et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med 22, 270–277 (2016). https://doi.org/10.1038/nm.4041
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4041
- Springer Nature America, Inc.
This article is cited by
-
Raltitrexed enhanced antitumor effect of anlotinib in human esophageal squamous carcinoma cells on proliferation, invasiveness, and apoptosis
BMC Cancer (2023)
-
A network map of discoidin domain receptor 1(DDR1)-mediated signaling in pathological conditions
Journal of Cell Communication and Signaling (2023)
-
Zebrafish patient-derived xenograft models predict lymph node involvement and treatment outcome in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
Discoidin Domain Receptor 2 orchestrates melanoma resistance combining phenotype switching and proliferation
Oncogene (2022)
-
DDR1 promotes hepatocellular carcinoma metastasis through recruiting PSD4 to ARF6
Oncogene (2022)