Abstract
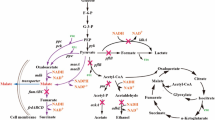
Deacidification of grape musts is crucial for the production of well-balanced wines, especially in colder regions of the world. The major acids in wine are tartaric and malic acid. Saccharomyces cerevisiae cannot degrade malic acid efficiently due to the lack of a malate transporter and the low substrate affinity of its malic enzyme. We have introduced efficient pathways for malate degradation in S. cerevisiae by cloning and expressing the Schizosaccharomyces pombe malate permease (mae1) gene with either the S. pombe malic enzyme (mae2) or Lactococcus lactis malolactic (mleS) gene in this yeast. Under aerobic conditions, the recombinant strain expressing the mae1 and mae2 genes efficiently degraded 8 g/L of malate in a glycerol-ethanol medium within 7 days. The recombinant malolactic strain of S. cerevisiae (mael and mleS genes) fermented 4.5 g/L of malate in a synthetic grape must within 4 days.
Similar content being viewed by others
References
Beelman, R.B. and Gallander, J.F. 1979. Wine deacidification. Adv. Food Res. 25: 1–53.
van Vuuren, H.J.J. and Dicks, L.M.T. Leuconostoc oenos: A review. Am. J. Enol. Vitic. 44: 99–112.
Davies, C.R., Wibowo, W., Eschenbruch, R., Lee, T.H., and Fleet, G.H. 1985. Practical implications of malolactic fermentation: A review. Am. J. Enol. Vitic. 36: 290–301.
Henick-Kling, T. 1995. Control of malolactic fermentation in wine: energetics, flavor modification and methods of starter culture preparation. J. Appl. Bacteriol. (Symp. Suppl.) 79: 29S–37S.
Gallander, J.F. 1977.Deacidification of Eastern table wines with Schizosaccharomyces pombe . Am. J. Enol. Vitic. 28: 65–68.
Radler, F. 1993. Yeast metabolism of organic acids pp. 165–182 in Wine Microbiology and Biotechnology. Fleet (ed.), Harwood Academic Publishers, Switzerland.
Gao, C. and Fleet, G.H. 1995. Degradation of malic and tartaric acids by high density cell suspensions of wine yeasts. Food Microbiol. 12: 65–71.
Mayer, K. and Temperli, A. 1963. The metabolism of L-malate and other compounds by Schizosaccharomyces pombe . Arch. Mikrobiol. 46: 321–328.
Fuck, E., Stark, G., and Radler, F. 1973. Äpfelsäurestoff-wechsel bei SaccharomycesII. Anreicherung und Eigenschaften eines Malatenzymes. Arch. Mikrobiol. 89: 223–231.
Temperli, A., Kunsch, V., Mayer, K., and Busch, I. 1965. Reinigung und Eigenschaften der Malatdehydrogenase (decarboxylierent) aus Hefe. Biochem. Biophys. Acta. 110: 630–632.
Ansanay, V., Dequin, S., Camarasa, C., Schaeffer, V., Grivet, J., Blondin, B., Salmon, J., and Barre, P. 1996. Malolactic fermentation by engineered Saccharomyces cerevisiaeas compared with engineered Schizosaccharomyces pombe . Yeast 12: 215–225.
Ansanay, V., Dequin, S., Blondin, B., and Barre, P., 1993. loning, sequence and expression of the gene encoding the malolactic enzyme from Lactococcus lactis . FEBS Lett. 332: 74–80.
Denayrolles, M., Aigle, M., and Lonvaud-Funel, A. 1995. Functional expression in Saccharomyces cerevisiaeof the Lactococcus lactis mleSgene encoding the malolactic enzyme. FEMS Microbiol. Lett. 125: 3744.
Williams, S.A., Hodges, R.A., Strike, T.L., Snow, R., and Kunkee, R.E. 1984. Cloning the gene for the malolactic fermentation of wine from Lactobacillus delbrueckiiin Escherichia coliand yeasts. Appl. Environ. Microbiol. 47: 288–293.
Grobler, J., Bauer, F., Subden, R.E., and van Vuuren, H.J.J. . 1995. The maelgene of Schizosaccharomyces pombeencodes a permease for malate and other C4 dicarboxylic acids. Veasf 11: 1485–1491.
Viljoen, M., Subden, R.E., Krizus, A., and van Vuuren, H.J.J. . 1994. Molecular analysis of the malic enzyme gene (mae2)of Schizosaccharomyces pombe . Yeast 10: 613–624.
Laing, E. and Pretorius, I.S. 1992. Synthesis and secretion of an Erwinia chrysanthemipectate lyase in Saccharomyces cerevisiaeregulated by different combinations of bacterial and yeast promoter and signal sequences. Gene 121: 35–45.
Maconi, E., Manachini, R., Aragozzini, F., Gennari, C., and Ricca, G. 1984. A study on the malo-alcoholic fermentation pathway in Schizosaccharomyces pombe . Biochem. J. 217: 585–588.
Murai, T., Tokushige, M., Nagai, J., and Katsuki, H. 1972. Studies on the regulatory functions of malic enzymes. J. Biochem. 71: 1015–1028.
Artus, N.N. and Edwards, G.E. 1985. NAD-malic enzyme from plants. FEBS Lett 182: 225–233.
Yanisch-Perron, C., Vieira, J., and Messing, J. 1985. Improved M13 cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33: 103–119.
Sambrook, J., Fritsh, E.F., and Maniatis, T. 1989. Molecular Cloning: A Laboratory Manual (2nd ed.). Cold Spring Harbor Laboratory Press, New York.
Sikorski, R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DMA in Saccharomyces cerevisiae . Genetics 122: 19–27.
Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (eds). 1995. Current protocols in molecular biology. John Wiley & Sons, New York.
Gietz, R.D. and Sugino, A., 1988. yeast-Escherichia colishuttle vectors constructed with in vitromutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534.
Kunkee, R.E. 1968. Simplified chromatographic procedure for detection of malolactic fermentation. Wines and Vines 49: 23–24.
Smith, D.B. and Johnson, K.S. 1988. Single step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene 67: 31–40.
Chirio, M.-C., Brethes, D., Napias, C., Grandier-Vazille, X., Rakotomanana, F., and Chevallier, J. 1990. Photoaffinity labelling of the purine-cytosine permease of Saccharomyces cerevisiae . Eur. J. Biochem. 194: 293–299.
Crous, J.M., Pretorius, I.S., and Van Zyl, W.H. 1995. Cloning and expression of an Aspergillus kawachiiendo-1,4-β-xylanase gene in Saccharomyces cerevisiae . Curr. Genet. 28: 467–473.
Martineau, B., Henick-Kling, T., and Acree, T. 1995. Reassessment of the influence of malolactic fermentation on the concentration of diacetyl in wines. Am. J. Enol. Vitic. 46: 385–388.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Volschenk, H., Viljoen, M., Grobler, J. et al. Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat Biotechnol 15, 253–257 (1997). https://doi.org/10.1038/nbt0397-253
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0397-253
- Springer Nature America, Inc.
This article is cited by
-
Yeast bioprospecting versus synthetic biology—which is better for innovative beverage fermentation?
Applied Microbiology and Biotechnology (2020)
-
Identification and engineering a C4-dicarboxylate transporter for improvement of malic acid production in Aspergillus niger
Applied Microbiology and Biotechnology (2020)
-
The use of lactic acid-producing, malic acid-producing, or malic acid-degrading yeast strains for acidity adjustment in the wine industry
Applied Microbiology and Biotechnology (2014)
-
Implications of new research and technologies for malolactic fermentation in wine
Applied Microbiology and Biotechnology (2014)
-
Surface Display of Malolactic Enzyme from Oenococcus oeni on Saccharomyces cerevisiae
Applied Biochemistry and Biotechnology (2013)