Abstract
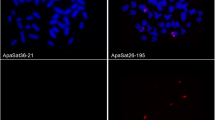
A highly repeated HpaI DNA family was revealed in Atlantic salmon (Salmo salar) and analyzed by Southern blotting and fluorescence in situ hybridization (FISH). In this report, we describe the nucleotide sequence, genomic structure and chromosomal localization of this HpaI repeat. This novel satellite appeared tandemly arrayed and located at centromeric areas of three acrocentric chromosome pairs as evidenced by FISH. The sequence was characterized by a high AT content (63%), a short consensus motif (A/T)(G/C)AAA(T/C) similar to other centromeric satellites motifs, and by short AT enriched stretches. The presence of this sequence in other salmonid species was also tested by Southern blot hybridization and used to analyze its evolution within this group.
Similar content being viewed by others
References
Abuín, M., C. Clabby, P. Martínez, U. Goswami, F. Flavin, N.P. Wilkins, J.A. Houghton, R. Powell & L. Sánchez, 1996a. A NOR-associated repetitive element present in the genome of two Salmo species (Salmo salar and Salmo trutta). Genome 39: 671–679.
Abuín, M., P. Martínez & L. Sánchez, 1996b. Localization of the repetitive telomeric sequence (TTAGGG)n in four salmonid species. Genome 39: 1035–1038.
Capriglione, T., A. Morescalchi, E. Olmo, L. Rocco, L. Stingo & S. Manzo, 1994. Satellite DNAs, heterochromatin and sex chromosomes in Chionodraco hamatus (Channichthyidae, Perciformes). Polar Biol. 14: 285–290.
Charlesworth, B., P. Sniegowski & W. Stephan, 1994. The evolutionary dynamics of repetititve DNA in eukaryotes. Nature 371: 215–220.
Cremisi, F., R. Vignali, R. Batistoni & G. Barsacchi, 1988. Heterochromatic DNA in Triturus (Amphibia, Urodela). II. A centromeric satellite DNA. Chromosoma 97: 204–211.
Choo, A.K.H., 2000. Centromerization. Trends Cell Biol. 10: 182–188.
Choo, A.K.H., 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1(2): 165–177.
Denovan, E.M. & J.M. Wright, 1990. A satellite DNA family from pollock (Pollachius virens). Gene 87: 279–283.
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Fischer, C., C. Ozouf-Costaz, H. Roest Crollius, C. Dasilva, O. Jaillon, L. Bouneau, C. Bonillo, J. Weissenbach & A. Bernot, 2000. Karyotype and chromosome location of characteristic tandem repeats in the pufferfish Tetraodon nigroviridis. Cytogenet. Cell Genet. 88: 50–55.
Garrido-Ramos, M.A., M. Jamilena, R. Lozano, C. Ruiz Rejó n & M. Ruiz Rejó n, 1995. The EcoRI centromeric satellite DNA of the Sparidae family (Pisces: Perciformes) contains a sequence motive common to other vertebrate centromeric satellite DNAs. Cytogenet. Cell Genet. 71: 345–351.
Goodier, J.L. & W.S. Davidson, 1998. Characterization of novel minisatellite repeat loci in Atlantic salmon (Salmo salar) and their phylogenetic distribution. J. Mol. Evol. 46: 245–255.
Grady, D.L., R.L. Ratliff, D.L. Robinson, E.C. McCanlies, J. Meyne & R.K. Moyzis, 1992. Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl. Acad. Sci. USA 89: 1695–1699.
Haaf, T., M. Schmid, C. Steinlein, P.M. Galetti & H. Willard, 1993. Organization and molecular cytogenetics of a satellite DNA family from Hoplias malabaricus (Pisces, Erythrinidae). Chromosome Res. 1: 77–86.
Harrington, J.J., G. Van Bokkelen, R.W. Mays, K. Gustashaw & H.F. Willard, 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15: 345–355.
Hartley, S.E. & W.S. Davidson, 1994. Distribution of satellite DNA sequences isolated from Arctic char, Salvelinus alpinus, in the genus Salvelinus. Can. J. Fish Aquat. Sci. 51(Suppl. 1): 277–283.
Hartley, S.E. & M.T. Horne 1984. Chromosome polymorphism and constitutive heterochromatin in Atlantic salmon, Salmo salar. Chromosoma 89: 377–380.
Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120.
Koehler, M.R., T. Haaf, M. Guttenbach, M. Schartl & M. Schmid, 1997. Cytogenetics of the genus Leporinus (Pisces, Anostomidae). II. Molecular cytogenetics, organization and evolutionary conservation of a chromosome-specific satellite DNA from Leporinus obtusidens. Chromosom Res. 5: 325–3331.
Kumar, S., K. Tamura, I.B. Jakobsen & M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17(2): 1244–1245.
Li, W.H. & D. Graur, 1991. Fundamentals of molecular evolution. Sinauer Associates Inc., Sunderland, MA, USA.
Martínez, P., A. Viñas, C. Bouza, J. Castro & L. Sánchez, 1993. Quantitative analysis of the variability of NOR region in Salmo trutta. Genome 36: 1119–1123.
Morán, P., K.M. Reed, J. Pérez, T.H. Oakley, R.B. Phillips, E. García-Vázquez, A. Pendás, 1997. Physical localization and characterization of the BglI element in the genomes of Atlantic salmon (Salmo salar) and brown trout (Salmo trutta L.). Gene 194: 9–18.
Murata, S., N. Takasaki, M. Saitoh, H. Tachida & N. Okada, 1996. Details of retropositional genome dynamics that provide a rationale for a generic division: the distinct branching of all the Pacific salmon and trout (Oncorhynchus) from the Atlantic salmon and trout (Salmo). Genetics 142: 915–926.
Ohno, S., 1970. Evolution by Gene Duplication. Springer, Berlin.
Ohno, S., 1972. So much 'junk' in our genomes. Brookhaven Symp. Biol. 23: 366–370.
Oliveira, C. & J.M. Wright, 1998. Molecular cytogenetic analysis of heterochromatin in the chromosomes of tilapia, Oreochromis niloticus (Teleostei: Cichlidae). Chromosome Res. 6: 205–211.
Orgel, L.E. & F.H.C. Crick, 1980. Selfish DNA: the ultimate parasite. Nature 284: 604–607.
Pendás, A.M., P. Morán & E. García-Vázquez, 1993. Ribosomal RNA genes are interspersed throughout a heterochromatic chromosome arm in Atlantic salmon. Cytogenet. Cell Genet. 63: 128–130.
Pérez, J., P. Morán & E. García-Vázquez, 2000. Isolation, characterization and chromosomal location of the tRNA(Met) genes in Atlantic salmon (Salmo salar) and brown trout (Salmo trutta). Genome 43(1): 185–190.
Phillips, R.B. & K.M. Reed, 1996. Application of fluorescence in situ hybridization (FISH) techniques to fish genetics: a review. Aquaculture 140: 197–216.
Reed, K.M. & R.B. Phillips, 1995. Molecular characterization and cytogenetic analysis of highly repeated DNAs of lake trout, Salvelinus namaycush. Chromosoma 104: 242–251.
Sambrook, J., E.F. Fritsch & T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Laboratory Press, Cold Spring Harbour.
Shepard, W., W.B.T. Cruse, R. Fourme, E. de la Fortelle & T. Prangé, 1998. A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 Å resolution. Structure 6: 849–861.
Thompson, J.D., D.G. Higgins & T.J. Gibson, 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 11(22): 4673–4680.
Vissel, B., A. Nagy & K.H.A. Choo, 1992. A satellite III sequence shared by human chromosomes 13, 14 and 21 that is contiguous with alpha satellite DNA. Cytogenet. Cell Genet. 61: 81–86.
Vogt, P., 1992. Code domains in tandem repeat DNA sequences structures. Chromosoma 101(10): 585–589.
Wong, A.K.C. & J.B. Rattner, 1988. Sequence organization and cytological localization of the minor satellite of mouse. Nucl. Acids Res. 16: 11645–11661.
Wright, J.M., 1989. Nucleotide sequence, genomic organization and evolution of a major repetitive DNA family in tilapia (Oreochromis mossambicus/hornorum). Nucl. Acids Res. 17: 5071–5079.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Viñas, A., Abuín, M., Pardo, B.G. et al. Characterization of a New HpaI Centromeric Satellite DNA in Salmo salar . Genetica 121, 81–87 (2004). https://doi.org/10.1023/B:GENE.0000019927.30049.9c
Issue Date:
DOI: https://doi.org/10.1023/B:GENE.0000019927.30049.9c