Abstract
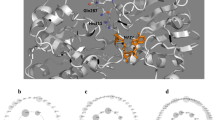
An homology model of Candida methylica formate dehydrogenase (cmFDH) was constructed based on the Pseudomonas sp. 101 formate dehydrogenase (psFDH) structure. In wild type cmFDH, Thr169 and Thr226 can form hydrogen bonds with each other. We measured the interaction energy between the two threonines independent of other interactions in the proteins by using a so-called double mutant cycle and assessing the protein stability from the concentration of guanidine hydrochloride needed to denature 50% of the molecules. We conclude that the hydrogen bonds stabilize the wild type protein by −4 kcal mol−1.
Similar content being viewed by others
References
Byrne MP, Manuel RL, LG Lowe, Stites WE (1995) Energetic contribution of side chain hydrogen bonding to the stability of staphylococcal nuclease. Biochemistry 34: 13949–13960.
Carter P, Winter G, Wilkinson A, Fersht AR (1984) The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell 38: 835–840. 1140
Fersht AR (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. NewYork: W.H. Freeman and Company.
Karaguler NG, Sessions RB, Clarke AR, Holbrook JJ (2001) A single mutation in the NAD-specific formate dehydrogenase from Candida methylicaallows the enzyme to use NADP. Biotechnol.Lett. 23: 283–287.
Karaguler NG, Sessions RB, Holbrook JJ (2001) Role of glutamate-52 in the mechanism of L-lactate dehydrogenase from Bacillus stearothermophilus. Biotechnol. Lett. 23: 395–399.
Lamzin V, Dauter Z, Popov V, Harutyunhan E, Wilson K (1994) High resolution structures of holo and apo formate dehydrogenase. J. Mol. Biol. 236: 759–785.
Lazaridis T, Archontis G, Karplus M (1995) Enthalpic contribution to protein stability: insights from atom-based calculations and statistical mechanics. Adv. Protein Chem. 47: 231–306.
Leatherbarrow RJ (1992) Grafit 3.0. Staines, UK: Erithacus Software.
Pace CN (1995) Evaluating contribution of hydrogen bonding and hydrophobic bonding to protein folding. Meth. Enzymol. 259: 538–554.
Serrano L, Horovitz A, Avron B, Bycroft M, Fersht AR (1990) Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double mutant cycles. Biochemistry 29: 9343–9352.
Sessions RB, Dewar V, Clarke AR, Holbrook JJ (1997) A model of plasmodium falciparumlactate dehydrogenase and its implications for design of improved antimalarials and the enhanced detection of parasitaemia. Protein Eng. 10: 301–306.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Karagüler, N.G., Sessions, R.B., Moreton, K.M. et al. Estimating the energetic contribution of hydrogen bonding to the stability of Candida methylica formate dehydrogenase by using double mutant cycle. Biotechnology Letters 26, 1137–1140 (2004). https://doi.org/10.1023/B:BILE.0000035485.43826.03
Issue Date:
DOI: https://doi.org/10.1023/B:BILE.0000035485.43826.03