Abstract
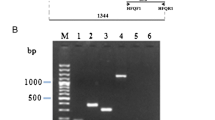
Bacillus subtilis aprE gene codes for the extracellular protease subtilisin. Its expression is controlled by AbrB, DegU, Hpr, SinI, SinR and Spo0A transition state protein regulators. To determine in vivo the protein–protein interactions among these regulators, we used the LexA-based bacterial genetic two-hybrid system. Our results show homo-dimerization to all the analyzed proteins and hetero-dimerization between SinR-SinI and SinR-Hpr.
Similar content being viewed by others
References
Bai U, Mandic-Mulec I, Smith I (1993) SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Develop. 7: 139–148.
Daines DA, Wright LF, Chaffin DO, Rubens CE, Silver RP (2000) NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol. Lett. 189: 281–284.
Dayle AD, Silver RP (2000) Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J. Bacteriol. 182: 5267–5270.
Dmitrova M, Younes-Cauet G, Oertel-Buchheit P, Porte D, Scnarr M, Granger S (1998) A new LexA-based genetic system for monitoring and analyzing protein heterodimeritation in Escherichia coli. Mol. Gen. Genet. 257: 205–212.
Ferrari E, Henner DJ, Perego M, Hoch J (1988) Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J. Bacteriol. 170: 289–295.
Fogh RH, Ottleben G, Ruterjans H, Schnarr M, Boelens R, Kaptein R (1994) Solution structure of the LexA represor DNA binding domain determined by H1NMR spectroscopy. EMBO J. 13: 3936–3944.
Gaur NK, Dubnau E, Smith I (1986) Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J. Bacteriol. 168: 860–869.
Gaur NK, Oppenheim J, Smith I (1991) The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173: 678–686.
Henner DJ, Ferrari E, Perego M, Hoch JA (1988) Location of the targets of the hpr-97, sacU32(Hy) and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J. Bacteriol. 170: 296–300.
Kallio PT, Fagelson JE, Hoch J, Strauch M (1991) The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266: 13411–13417.
Kunst F, Debarbouille M, Msadek T, Young M, Mauel C, Karamata D, Klier A, Rapoport G, Dedonder R (1988) Deduced polypeptides encoded by the Bacillus subtilis sacU locus share homology with two-component sensor-regulator systems. J. Bacteriol. 170: 5093–5101.
Kunst F, Pascal M, Lepesant KJ, Lepesant J, Billault A, Dedonder R (1974) Pleiotropic mutations affecting sporulation conditions and the synthesis of extracellular enzymes in Bacillus subtilis 168. Biochimie 56: 1481–1489.
Lamerich R, Padilla A, Boelens R, Kaptein R, Ottleben G, Ruterjans H, Granger S, Oertel P, Schnarr M (1989) The amino-terminal domain of LexA repressor is a a-helical but differs from canonical helix-turn-helix proteins: a two dimensional study. Proc. Natl. Acad. Sci. USA 86: 6863–6867.
Lewis RJ, Bannigan JA, Smith I, Wilkinson AJ (1996) Crystallisation of the Bacillus subtilis sporulation inhibitor SinR, complexed with its antagonist, SinI. FEBS Lett. 378: 98–100.
Mandic-Mulec I, Doukhan L, Smith I (1995) The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spoOA. J. Bacteriol. 19: 4619–4627.
Olmos SJ, Contreras FR (2003) Genetic system constructed to overproduce and secrete proinsulin in Bacillus subtilis. Appl. Microbiol. Biotechnol. 62: 369–373.
Olmos SJ, Bolaños V, Causey S, Ferrari E, Bolivar F, Valle F (1996) A functional Spo0A is required for maximal aprE expression in Bacillus subtilis. FEBS Lett. 381: 29–31.
Olmos SJ, DeAnda R, Ferrari E, Bolivar F, Valle F (1997) Effects of the sinR and degU32 (Hy) mutations on the regulation of the aprE gene in Bacillus subtilis. Mol. Gen. Genet. 253: 562–567
Olmos J, Sanchez A, De Anda R (1998) Regulation of the aprE (subtilisin) gene in abrB mutants of Bacillus subtilis. Asia-Pac. J. Mol. Biol. Biotechnol. 6: 97–103.
Perego M, Wu JJ, Spiegelman GB, Hoch JA (1991) Mutational dissociation of the positive and negative regulatory properties of the Spo0A sporulation transcription factor of Bacillus subtilis. Gene 100: 207–212.
Porte D, Oertel-Buchheit P, Granger S, Schnarr M (1995) Fos leucine zipper variants with increased association capacity. J. Biol. Chem. 29: 22721–22730.
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schnarr M, Granger SM (1993) LexA, the self-cleaving transcriptional repressor of the SOS system. Nucl. Acids Mol. Biol. 7: 170–189.
Sonenshein AL, Losick R, Hoch JA (1993) Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology and Molecular Genetics. Washington, DC: American Society for Microbiology, pp. 987.
Strauch MA, Hoch JA (1995) Control of postesponential gene expression by transition state regulators. In: Doi HR, McGloughlin M, eds. Biology of Bacilli: Applications to Industry. Boston MA: Butterworth-Heineman, pp. 105–121.
Strauch MA, Perego M, Burbulys D, Hoch JA (1989) The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol. Microbiol. 3: 1203–1209.
Wong SL (1995) Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr. Opin. Biotechnol. 6: 517–522.
Xu K, Strauch M (2001) DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J. Bacteriol. 183: 4094–4098.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sánchez, A., Olmos, J. Bacillus subtilis transcriptional regulators interaction. Biotechnology Letters 26, 403–407 (2004). https://doi.org/10.1023/B:BILE.0000018259.66762.ed
Issue Date:
DOI: https://doi.org/10.1023/B:BILE.0000018259.66762.ed