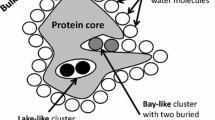
Abstract
We have studied the classification of the environment of residues within protein structures. Eisenberg's original idea created environmental categories to discriminate between similar residues [Bowie et al., Science (1991), 253, 164–170]. These environments grouped residues based upon their buried surface area, polarity of the surrounding environment, and secondary structure element in which the residue is found. However, Eisenberg's original categories led to incomplete discrimination between residues that only partially substitute for each other. We have expanded on Eisenberg's original idea of environmental categories, by both considering additional contacts in the calculation of the solvent-accessible molecular surface area and by subdividing the environmental plot into regions based upon its theoretical features. Our alternative surface area calculations were used in conjunction with the polarity of the environment of the residue to define a new set of environmental categories. These new categories were able to discriminate between residues such as threonine, valine, and aspartic acid while reflecting the propensity of these residues to substitute for each other.
Similar content being viewed by others
REFERENCES
Altschul, S. F., Carroll, R. J., and Lipman, D. J. (1989). Weights for data related by a tree, J. Mol. Biol. 207, 647–653.
Bowie, J. U., and Eisenberg, D. (1993). Inverted protein structure prediction, Curr. Opin. Struct. Biol. 3, 437–444.
Bowie, J. U., and Eisenberg, D. (1994). An evolutionary approach to folding small α-helical proteins that uses sequence information and an empirical guiding fitness function, Proc. Natl. Acad. Sci. USA 91, 4436–4440.
Bowie, J. U., Reidhaar-Olson, J. F., Lim, W. A., and Sauer, R. T. (1990). Deciphering the message in protein sequences: Tolerance to amino acid substitutions, Science 247, 1306–1310.
Bowie, J. U., Lüthy, R., and Eisenberg, D. (1991). A method to identify protein sequences that fold into a known three-dimensional structure, Science 253, 164–170.
Bowie, J. U., Lüthy, R., and Eisenberg, D. (1995). Three-dimensional profiles: Assessing the compatibility of an amino acid sequence with a three-dimensional structure, in Tracing Biological Evolution in Protein and Gene Structures (Go, M., and Schimmel, P., eds.), Elsevier Science, pp. 297–309.
Bryant, S. H., and Amzel, L. M. (1987). Correctly folded proteins make twice as many hydrophobic contacts, Int. J. Peptide Res. 29, 46–52.
Collins, J. F., and Coulson, F. W. (1990). Significance of protein sequence similarities, Meth. Enzymol. 183, 474–487.
Connolly, M. L. (1983). Analytical molecular surface calculation, J. Appl. Crystallogr., 16, 548–558.
Connolly, M. L. (1985a). J. Am. Chem. Soc. 107, 1118–1124.
Connolly, M. L. (1985b). J. Appl. Crystallogr. 18, 499–505.
Dill, K. A. (1990). Dominant forces in protein folding, Biochemistry 29, 7133–7155.
Drexler, K. E. (1981). Molecular engineering: an approach to the development of general capabilities for molecular manipulation, Proc. Natl. Acad. Sci. USA 78, 5275–5278.
Engh, R. A., and Huber, R. (1991). Accurate bond angle parameters for X-ray protein structure refinement, Acta Crystallogr. A 47, 392–400.
Francl, M. M., Hunt, Jr., R. F., and Hehre, W. J. (1984). J. Am. Chem. Soc. 106, 563–570.
Johnson, M. S., and Overington, J. P. (1993). A structural basis for sequence comparisons. An evaluation of scoring methodologies, J. Mol. Biol. 233, 716–738.
Jordan, J. (1973). J. Mol. Biol., 41, 375–395.
Kabsch, W., and Sander, C. (1983). Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features, Biopolymers 22, 2577–2637.
Kauzmann, W. (1959). Adv. Protein Chem. 14, 1.
Lee, B., and Richards, F. M. (1971). The interpretation of protein structures: Estimation of static accessibility, J. Mol. Biol. 55, 379–400.
Lüthy, R., Bowie, J. U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles, Nature 356, 83–85.
Pabo, C. (1983). Designing proteins and peptides. Nature 301, 200.
Pauling, L. (1960). The Nature of the Chemical Bond, Cornell University Press, Ithaca, New York, p. 257.
Reidhaar-Olson, J. F., and Sauer, R. T. (1988). Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences, Science 241, 53–57.
Sanders, S., and Kabisch, W. (1983). Biopolymers 22, 2577–2637.
Sellers, P. H. (1980). On the theory and computation of evolutionary distances: Pattern recognition, J. Algorithms 1, 359–373.
Sweet, R. M., and Eisenberg, D. (1983). Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure, J. Mol. Biol. 171, 479–488.
Wilmanns, M., and Eisenberg, D. (1993). Three-dimensional profiles from residue-pair preferences: Identification of sequence with β/α-barrel fold, Proc. Natl. Acad. Sci. USA 90, 1379–1383.
Zhang, K. Y. J., and Eisenberg, D. (1994). The three-dimensional profile method using residue preference as a continuous function of residue environment, Protein Sci. 3, 687–695.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Deerfield, D.W., Holland-Minkley, A.M., Geigel, J. et al. Classification of the Environment of Protein Residues. J Protein Chem 16, 441–447 (1997). https://doi.org/10.1023/A:1026349124850
Published:
Issue Date:
DOI: https://doi.org/10.1023/A:1026349124850